An abdominal obesity missense variant in the adipocyte thermogenesis gene TBX15 is implicated in adaptation to cold in Finns
- PMID: 39515300
- PMCID: PMC11568758
- DOI: 10.1016/j.ajhg.2024.10.001
An abdominal obesity missense variant in the adipocyte thermogenesis gene TBX15 is implicated in adaptation to cold in Finns
Abstract
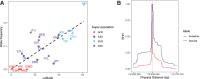
Mechanisms of abdominal obesity GWAS variants have remained largely unknown. To elucidate these mechanisms, we leveraged subcutaneous adipose tissue (SAT) single nucleus RNA-sequencing and genomics data. After discovering that heritability of abdominal obesity is enriched in adipocytes, we focused on a SAT unique adipocyte marker gene, the transcription factor TBX15, and its abdominal obesity-associated deleterious missense variant, rs10494217. The allele frequency of rs10494217 revealed a north-to-south decreasing gradient, with consistent significant FST values observed for 25 different populations when compared to Finns, a population with a history of genetic isolation. Given the role of Tbx15 in mouse thermogenesis, the frequency may have increased as an adaptation to cold in Finns. Our selection analysis provided significant evidence of selection for the abdominal obesity risk allele T of rs10494217 in Finns, with a north-to-south decreasing trend in other populations, and demonstrated that latitude significantly predicts the allele frequency. We also discovered that the risk allele status significantly affects SAT adipocyte expression of multiple adipocyte marker genes in trans in two cohorts. Two of these trans genes have been connected to thermogenesis, supporting the thermogenic effect of the TBX15 missense variant as a possible cause of its selection. Adipose expression of one trans gene, a lncRNA, AC002066.1, was strongly associated with adipocyte size, implicating it in metabolically unhealthy adipocyte hypertrophy. In summary, the abdominal obesity variant rs10494217 was selected in Finns, and individuals with the risk allele have trans effects on adipocyte expression of genes relating to thermogenesis and adipocyte hypertrophy.
Keywords: SAT; T-box transcription factor 15; TBX15; WHRadjBMI; abdominal obesity; adipocyte hypertrophy; population genetics; selection; single nucleus RNA sequencing; subcutaneous adipose tissue; thermogenesis; trans regulation; waist-hip ratio adjusted for body mass index.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures



References
-
- Martin S.S., Aday A.W., Almarzooq Z.I., Anderson C.A.M., Arora P., Avery C.L., Baker-Smith C.M., Barone Gibbs B., Beaton A.Z., Boehme A.K., et al. 2024 Heart Disease and Stroke Statistics: A Report of US and Global Data From the American Heart Association. Circulation. 2024;149:e347–e913. doi: 10.1161/CIR.0000000000001209. - DOI - PubMed
-
- Seidell J.C., Björntorp P., Sjöström L., Sannerstedt R., Krotkiewski M., Kvist H. Regional distribution of muscle and fat mass in men--new insight into the risk of abdominal obesity using computed tomography. Int. J. Obes. 1989;13:289–303. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

