Comparative analysis of the postadmission and antemortem oropharyngeal and rectal swab microbiota of ICU patients
- PMID: 39516251
- PMCID: PMC11549221
- DOI: 10.1038/s41598-024-78102-1
Comparative analysis of the postadmission and antemortem oropharyngeal and rectal swab microbiota of ICU patients
Abstract
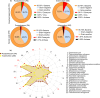
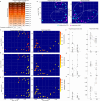
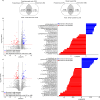
Shotgun metabarcoding was conducted to examine the microbiota in a total of 48 samples from 12 critically ill patients, analyzing samples from both the oropharynx and rectum. We aimed to compare their postadmission microbiota, characterized as moderately dysbiotic, with the severely dysbiotic antemortem microbiota associated with patients' deaths. We found that, compared with postadmission samples, patient antemortem swab samples presented moderate but not significantly decreased diversity indices. The antemortem oropharyngeal samples presented an increase in biofilm-forming bacteria, including Streptococcus oralis, methicillin-resistant Staphylococcus aureus (MRSA), and Enterococcus faecalis. Although the septic shock rate was 67%, no significant differences were detected in the potential pathogen ratios when the microbiota was analyzed. A notable strain-sharing rate between the oropharynx and intestine was noted. By comparing postadmission and antemortem samples, microbial biomarkers of severe dysbiosis were pinpointed through the analysis of differentially abundant and uniquely emerging species in both oropharyngeal and rectal swabs. Demonstrating strong interconnectivity along the oral-intestinal axis, these biomarkers could serve as indicators of the progression of dysbiosis. Furthermore, the microbial networks of the oropharyngeal microbiota in deceased patients presented the lowest modularity, suggesting a vulnerable community structure. Our data also highlight the critical importance of introducing treatments aimed at enhancing the resilience of the oral cavity microbiome, thereby contributing to better patient outcomes.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

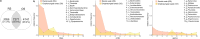

Similar articles
-
Rectal Swabs from Critically Ill Patients Provide Discordant Representations of the Gut Microbiome Compared to Stool Samples.mSphere. 2019 Jul 24;4(4):e00358-19. doi: 10.1128/mSphere.00358-19. mSphere. 2019. PMID: 31341070 Free PMC article.
-
Upper Respiratory Dysbiosis with a Facultative-dominated Ecotype in Advanced Lung Disease and Dynamic Change after Lung Transplant.Ann Am Thorac Soc. 2019 Nov;16(11):1383-1391. doi: 10.1513/AnnalsATS.201904-299OC. Ann Am Thorac Soc. 2019. PMID: 31415219 Free PMC article.
-
Oropharyngeal microbial ecosystem perturbations influence the risk for acute respiratory infections in common variable immunodeficiency.Front Immunol. 2024 May 30;15:1371118. doi: 10.3389/fimmu.2024.1371118. eCollection 2024. Front Immunol. 2024. PMID: 38873612 Free PMC article.
-
Links between oropharyngeal microbiota and IgA nephropathy: A paradigm shift from isolated microbe to microbiome.Microbiol Res. 2025 Mar;292:128005. doi: 10.1016/j.micres.2024.128005. Epub 2024 Dec 8. Microbiol Res. 2025. PMID: 39675141 Review.
-
Pathogenesis and therapeutic opportunities of gut microbiome dysbiosis in critical illness.Gut Microbes. 2024 Jan-Dec;16(1):2351478. doi: 10.1080/19490976.2024.2351478. Epub 2024 May 23. Gut Microbes. 2024. PMID: 38780485 Free PMC article. Review.
Cited by
-
Analysis of ICU resistome dynamics in patients, staff and environment for the identification of predictive biomarkers of sepsis and early mortality.Sci Rep. 2025 Jul 11;15(1):25080. doi: 10.1038/s41598-025-10848-8. Sci Rep. 2025. PMID: 40646273 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

