CARD16 restores tumorigenesis and restraints apoptosis in glioma cells Via FOXO1/TRAIL axis
- PMID: 39516471
- PMCID: PMC11549220
- DOI: 10.1038/s41419-024-07196-2
CARD16 restores tumorigenesis and restraints apoptosis in glioma cells Via FOXO1/TRAIL axis
Abstract
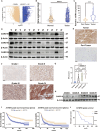
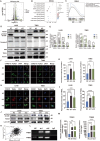
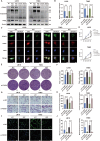
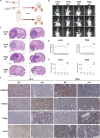
A hallmark of glioma cells, particularly glioblastoma multiforme (GBM) cells, is their resistance to apoptosis. Accumulating evidences has demonstrated that CARD16, a caspase recruitment domain (CARD) only protein, enhances both anti-apoptotic and tumorigenic properties. Nevertheless, there is a limited understanding of the expression and functional role of CARD16 in glioma. This study seeks to investigate, through in silico analysis and clinical specimens, the role of CARD16 as a potential tumor promoter in glioma. Functional assays and molecular studies revealed that CARD16 promotes tumorigenesis and suppresses apoptosis in glioma cells. Moreover, knockdown of CARD16 enhances the expression of the FOXO1/TRAIL axis in GBM cells. Additionally, FOXO1 downregulation in CARD16 knockdown GBM cells restores proliferation and reduces apoptosis. Further investigation demonstrated that elevated P21 expression inhibits CDK2-mediated FOXO1 phosphorylation and ubiquitination in CARD16-knockdown GBM cells. Collectively, these findings suggest that CARD16 is a tumor-promoting molecular in glioma via downregulating FOXO1/TRAIL axis, and suppressing TRAIL-induced apoptosis. The CARD16 gene presents significant potential for prognostic prediction and advances in innovative apoptotic therapeutics.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Lapointe S, Perry A, Butowski NA. Primary brain tumours in adults. Lancet. 2018;392:432–46. - PubMed
-
- Tan AC, Ashley DM, Lopez GY, Malinzak M, Friedman HS, Khasraw M. Management of glioblastoma: State of the art and future directions. CA Cancer J Clin. 2020;70:299–312. - PubMed
-
- Newton K, Strasser A, Kayagaki N, Dixit VM. Cell death. Cell. 2024;187:235–56. - PubMed
-
- Yuan J, Ofengeim D. A guide to cell death pathways. Nat Rev Mol Cell Biol. 2024;25:379–95. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

