ZBTB18 regulates cytokine expression and affects microglia/macrophage recruitment and commitment in glioblastoma
- PMID: 39516530
- PMCID: PMC11549471
- DOI: 10.1038/s42003-024-07144-y
ZBTB18 regulates cytokine expression and affects microglia/macrophage recruitment and commitment in glioblastoma
Abstract
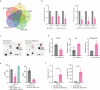
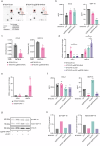
Glioma associated macrophages/microglia (GAMs) play an important role in glioblastoma (GBM) progression, due to their massive recruitment to the tumor site and polarization to a tumor promoting phenotype. GAMs secrete a variety of cytokines, which facilitate tumor cell growth and invasion, and prevent other immune cells from mounting an immune response against the tumor. Here, we demonstrate that zinc finger and BTB containing domain 18 (ZBTB18), a transcriptional repressor with tumor suppressive function in glioblastoma, impairs the production of key cytokines, which function as chemoattractant for GAMs. Consistently, we observe a reduced migration of GAMs when ZBTB18 is expressed by glioblastoma cells, both in cell culture and in vivo experiments. Moreover, RNA sequencing analysis shows that the presence of ZBTB18 in glioblastoma cells alters the commitment of conditioned microglia, suggesting the loss of the immune-suppressive phenotype and the acquisition of pro-inflammatory features. Thus, therapeutic approaches to increase ZBTB18 expression in GBM cells could represent an effective adjuvant to immune therapy in GBM.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






References
-
- Phillips, H. S. et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell9, 157–173 (2006). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

