Association analysis of gut microbiota with LDL-C metabolism and microbial pathogenicity in colorectal cancer patients
- PMID: 39516755
- PMCID: PMC11546423
- DOI: 10.1186/s12944-024-02333-4
Association analysis of gut microbiota with LDL-C metabolism and microbial pathogenicity in colorectal cancer patients
Abstract
Background: Colorectal cancer (CRC) is the most common gastrointestinal malignancy worldwide, with obesity-induced lipid metabolism disorders playing a crucial role in its progression. A complex connection exists between gut microbiota and the development of intestinal tumors through the microbiota metabolite pathway. Metabolic disorders frequently alter the gut microbiome, impairing immune and cellular functions and hastening cancer progression.
Methods: This study thoroughly examined the gut microbiota through 16S rRNA sequencing of fecal samples from 181 CRC patients, integrating preoperative Low-density lipoprotein cholesterol (LDL-C) levels and RNA sequencing data. The study includes a comparison of microbial diversity, differential microbiological analysis, exploration of the associations between microbiota, tumor microenvironment immune cells, and immune genes, enrichment analysis of potential biological functions of microbe-related host genes, and the prediction of LDL-C status through microorganisms.
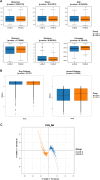
Results: The analysis revealed that differences in α and β diversity indices of intestinal microbiota in CRC patients were not statistically significant across different LDL-C metabolic states. Patients exhibited varying LDL-C metabolic conditions, leading to a bifurcation of their gut microbiota into two distinct clusters. Patients with LDL-C metabolic irregularities had higher concentrations of twelve gut microbiota, which were linked to various immune cells and immune-related genes, influencing tumor immunity. Under normal LDL-C metabolic conditions, the protective microorganism Anaerostipes_caccae was significantly negatively correlated with the GO Biological Process pathway involved in the negative regulation of the unfolded protein response in the endoplasmic reticulum. Both XGBoost and MLP models, developed using differential gut microbiota, could forecast LDL-C levels in CRC patients biologically.
Conclusions: The intestinal microbiota in CRC patients influences the LDL-C metabolic status. With elevated LDL-C levels, gut microbiota can regulate the function of immune cells and gene expression within the tumor microenvironment, affecting cancer-related pathways and promoting CRC progression. LDL-C and its associated gut microbiota could provide non-invasive markers for clinical evaluation and treatment of CRC patients.
Keywords: 16S rRNA; Clinical status; Colorectal cancer (CRC); Gut microbiota; LDL-C; Machine learning.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71(3):209–49. - PubMed
-
- Siegel RL, Miller KD, Goding Sauer A, Fedewa SA, Butterly LF, Anderson JC, Cercek A, Smith RA, Jemal A. Colorectal cancer statistics, 2020. CA Cancer J Clin. 2020;70(3):145–64. - PubMed
-
- Després JP, Lemieux I. Abdominal obesity and metabolic syndrome. Nature. 2006;444(7121):881–7. - PubMed
MeSH terms
Substances
Grants and funding
- yuanqingji2023-10hao/Youth Research Project of Guangxi Medical University Affiliated Cancer Hospital
- GXMUYSF202402/Youth Science Foundation of Guangxi Medical University
- GXMUYSF202357/Youth Science Foundation of Guangxi Medical University
- 2021GXNSFAA196008/Natural Science Foundation of Guangxi Province
- 2021KY0087/Middle-aged and Young Teachers' Basic Ability Promotion Project of Guangxi
LinkOut - more resources
Full Text Sources
Medical

