Exploring multi-omics and clinical characteristics linked to accelerated biological aging in Asian women of reproductive age: insights from the S-PRESTO study
- PMID: 39516835
- PMCID: PMC11549770
- DOI: 10.1186/s13073-024-01403-7
Exploring multi-omics and clinical characteristics linked to accelerated biological aging in Asian women of reproductive age: insights from the S-PRESTO study
Abstract
Background: Phenotypic age (PhenoAge), a widely used marker of biological aging, has been shown to be a robust predictor of all-cause mortality and morbidity in different populations. Existing studies on biological aging have primarily focused on individual domains, resulting in a lack of a comprehensive understanding of the multi-systemic dysregulation that occurs in aging.
Methods: PhenoAge was evaluated based on a linear combination of chronological age (CA) and 9 clinical biomarkers in 952 multi-ethnic Asian women of reproductive age. Phenotypic age acceleration (PhenoAgeAccel), an aging biomarker, represents PhenoAge after adjusting for CA. This study conducts an in-depth association analysis of PhenoAgeAccel with clinical, nutritional, lipidomic, gut microbiome, and genetic factors.
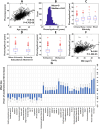
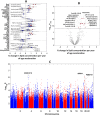
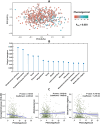
Results: Higher adiposity, glycaemia, plasma saturated fatty acids, kynurenine pathway metabolites, GlycA, riboflavin, nicotinamide, and insulin-like growth factor binding proteins were positively associated with PhenoAgeAccel. Conversely, a healthier diet and higher levels of pyridoxal phosphate, all-trans retinol, betaine, tryptophan, glutamine, histidine, apolipoprotein B, and insulin-like growth factors were inversely associated with PhenoAgeAccel. Lipidomic analysis found 132 lipid species linked to PhenoAgeAccel, with PC(O-36:0) showing the strongest positive association and CE(24:5) demonstrating the strongest inverse association. A genome-wide association study identified rs9864994 as the top genetic variant (P = 5.69E-07) from the ZDHHC19 gene. Gut microbiome analysis revealed that Erysipelotrichaceae UCG-003 and Bacteroides vulgatus were inversely associated with PhenoAgeAccel. Integrative network analysis of aging-related factors underscored the intricate links among clinical, nutritional and lipidomic variables, such as positive associations between kynurenine pathway metabolites, amino acids, adiposity, and insulin resistance. Furthermore, potential mediation effects of blood biomarkers related to inflammation, immune response, and nutritional and energy metabolism were observed in the associations of diet, adiposity, genetic variants, and gut microbial species with PhenoAgeAccel.
Conclusions: Our findings provide a comprehensive analysis of aging-related factors across multiple platforms, delineating their complex interconnections. This study is the first to report novel signatures in lipidomics, gut microbiome and blood biomarkers specifically associated with PhenoAgeAccel. These insights are invaluable in understanding the molecular and metabolic mechanisms underlying biological aging and shed light on potential interventions to mitigate accelerated biological aging by targeting modifiable factors.
Keywords: Age acceleration; Biological aging; GWAS; Gut microbiome; Lipidomics; PhenoAge.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

