Molecular Cloning, Characterization, and Application of a Novel Multifunctional Isoamylase (MIsA) from Myxococcus sp. Strain V11
- PMID: 39517265
- PMCID: PMC11544908
- DOI: 10.3390/foods13213481
Molecular Cloning, Characterization, and Application of a Novel Multifunctional Isoamylase (MIsA) from Myxococcus sp. Strain V11
Abstract
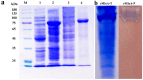
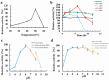
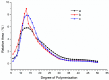
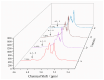
A novel multifunctional isoamylase, MIsA from Myxococcus sp. strain V11, was expressed in Escherichia coli BL21(DE3). Sequence alignment revealed that MIsA is a typical isoamylase that belongs to glycoside hydrolase family 13 (GH 13). MIsA can hydrolyze the α-1,6-branches of amylopectin and pullulan, as well as the α-1,4-glucosidic bond in amylose. Additionally, MIsA demonstrates 4-α-D-glucan transferase activity, enabling the transfer of α-1,4-glucan oligosaccharides between molecules, particularly with linear maltooligosaccharides. The Km, Kcat, and Vmax values of the MIsA for amylopectin were 1.22 mM, 40.42 µmol·min-1·mg-1, and 4046.31 mM·min-1. The yields of amylopectin and amylose hydrolyzed into oligosaccharides were 10.16% and 11.70%, respectively. The hydrolysis efficiencies were 55%, 35%, and 30% for amylopectin, soluble starch, and amylose, respectively. In the composite enzyme hydrolysis of amylose, the yield of maltotetraose increased by 1.81-fold and 2.73-fold compared with that of MIsA and MTHase (MCK8499120) alone, respectively.
Keywords: 4-alpha-glucanotransferase; Myxococcus sp. strain V11; isoamylase; maltotetraose; multifunctional starch debranching enzyme.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






Similar articles
-
TreX from Sulfolobus solfataricus ATCC 35092 displays isoamylase and 4-alpha-glucanotransferase activities.Biosci Biotechnol Biochem. 2007 May;71(5):1348-52. doi: 10.1271/bbb.70016. Epub 2007 May 7. Biosci Biotechnol Biochem. 2007. PMID: 17485831
-
Activity, cloning, and expression of an isoamylase-type starch-debranching enzyme from banana fruit.J Agric Food Chem. 2004 Dec 1;52(24):7412-8. doi: 10.1021/jf049300g. J Agric Food Chem. 2004. PMID: 15563228
-
Enzymatic synthesis and properties of highly branched rice starch amylose and amylopectin cluster.J Agric Food Chem. 2008 Jan 9;56(1):126-31. doi: 10.1021/jf072508s. Epub 2007 Dec 12. J Agric Food Chem. 2008. PMID: 18072737
-
Starch modification with microbial alpha-glucanotransferase enzymes.Carbohydr Polym. 2013 Mar 1;93(1):116-21. doi: 10.1016/j.carbpol.2012.01.065. Epub 2012 Jan 31. Carbohydr Polym. 2013. PMID: 23465909 Review.
-
Towards a better understanding of the metabolic system for amylopectin biosynthesis in plants: rice endosperm as a model tissue.Plant Cell Physiol. 2002 Jul;43(7):718-25. doi: 10.1093/pcp/pcf091. Plant Cell Physiol. 2002. PMID: 12154134 Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

