Diagnosis of Pancreatic Ductal Adenocarcinoma Using Deep Learning
- PMID: 39517902
- PMCID: PMC11548667
- DOI: 10.3390/s24217005
Diagnosis of Pancreatic Ductal Adenocarcinoma Using Deep Learning
Abstract
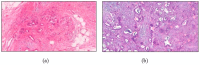
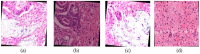
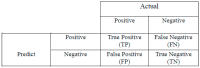
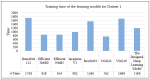
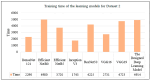
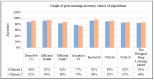
Recent advances in artificial intelligence (AI) research, particularly in image processing technologies, have shown promising applications across various domains, including health care. There is a significant effort to use AI for the early diagnosis and detection of diseases, offering cost-effective and timely solutions to enhance patient outcomes. This study introduces a deep learning network aimed at analyzing pathology images for the accurate diagnosis of pancreatic cancer, specifically pancreatic ductal adenocarcinoma (PDAC). Utilizing a novel dataset comprised of cases diagnosed with PDAC and/or chronic pancreatitis, this study applies deep learning algorithms to assess the effectiveness and reliability of the diagnostic process. The dataset was enhanced through image duplication and the creation of a second dataset with varied dimensions, facilitating the training of advanced transfer learning models including InceptionV3, DenseNet, ResNet, VGG, EfficientNet, and a specially designed deep neural network. The study presents a convolutional neural network model, optimized for the rapid and accurate detection of pancreatic cancer, and conducts a comparative analysis with other models to select the most accurate algorithm for a decision support system. The results from Dataset 1 show that EfficientNetB0 achieved a high success rate of 92%. In Dataset 2, VGG16 was found to have high performance, with a success rate of 92%. On the other hand, ResNet50 achieved a remarkable success rate of 96% despite a moderate training time and showed high precision, recall, F1 score, and accuracy. These results provide valuable data to demonstrate and share the relevance of different deep learning models in pancreatic cancer diagnosis.
Keywords: classification; convolutional neural networks; deep learning; health services; pancreatic ductal adenocarcinoma; pathology images.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
-
- Ahmadvand P., Farahani H., Farnell D., Darbandsari A., Topham J., Karasinska J., Nelson J., Naso J., Jones S.J.M., Renouf D., et al. A Deep Learning Approach for the Identification of the Molecular Subtypes of Pancreatic Ductal Adenocarcinoma Based on Whole Slide Pathology Images. Am. J. Pathol. 2024 doi: 10.1016/j.ajpath.2024.08.006. - DOI - PubMed
-
- Kriegsmann M., Kriegsmann K., Steinbuss G., Zgorzelski C., Kraft A., Gaida M.M. Deep learning in pancreatic tissue: Dentification of anatomical structures, pancreatic intraepithelial neoplasia, and ductal adenocarcinoma. Int. J. Mol. Sci. 2021;22:5385. doi: 10.3390/ijms22105385. - DOI - PMC - PubMed
-
- Deepthi G., Bamini A.A., Praveen Y.J. A Comparative Analysis of Pancreatic Tumor Detection using VGG16, ResNet, and DenseNet; Proceedings of the 2024 IEEE 3rd International Conference on Applied Artificial Intelligence and Computing (ICAAIC); Salem, India. 5–7 June 2024; pp. 1303–1308.
-
- Kronberg R.M., Haeberle L., Pfaus M., Xu H.C., Krings K.S., Schlensog M., Rau T., Pandyra A.A., Lang K.S., Esposito I., et al. Communicator-driven data preprocessing improves deep transfer learning of histopathological prediction of pancreatic ductal adenocarcinoma. Cancers. 2022;14:1964. doi: 10.3390/cancers14081964. - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

