The Evidence Base for Circulating Tumor DNA-Methylation in Non-Small Cell Lung Cancer: A Systematic Review and Meta-Analysis
- PMID: 39518079
- PMCID: PMC11544801
- DOI: 10.3390/cancers16213641
The Evidence Base for Circulating Tumor DNA-Methylation in Non-Small Cell Lung Cancer: A Systematic Review and Meta-Analysis
Abstract
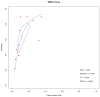
Background: Non-Small Cell Lung Cancer (NSCLC) remains a challenging disease to manage with effectiveness. Early detection and precise monitoring are crucial for improving patient outcomes. Circulating tumor DNA (ctDNA) offers a non-invasive cancer detection and monitoring method. Emerging biomarkers, such as ctDNA methylation, have shown promise in enhancing diagnostic accuracy and prognostic assessment in NSCLC. In this review, we examined the current evidence regarding ctDNA methylation's role in NSCLC detection through a systematic review of the existing literature and meta-analysis. Methods: We systematically searched PubMed, Medline, Embase, and Web of Science databases up to 26 June 2024 for studies on the role of ctDNA methylation analysis in NSCLC patients. We included studies from 2010 to 2024 on NSCLC patients. We excluded case reports, non-English articles, studies on cell lines or artificial samples, those without cfDNA detection, prognostic studies, and studies with non-extractable data or mixed cancer types. Funnel plots were visually examined for potential publication bias, with a p value < 0.05 indicating bias. Meta-analysis was conducted using R packages (meta, forestplot, and mada). Combined sensitivity, specificity, positive likelihood ratio (LR+), negative likelihood ratio (LR-), positive and negative predictive values, diagnostic odds ratio (DOR), and 95% confidence intervals (95% CI) were calculated. A summary receiver operating characteristic curve (SROC) and area under the curve (AUC) with related Standard Error (SE) were used to evaluate the overall diagnostic performance. Additionally, RASSF1A, APC, SOX17, SEPT9, and RARβ2 were analyzed, since their methylation was assessed in two or more studies. Results: From 38 candidate papers, we finally identified 12 studies, including 472 NSCLC patients. The pooled sensitivity was 0.62 (0.47-0.77) and the specificity was 0.90 (0.85-0.94). The diagnostic odds ratio was 15.6 (95% CI 9.36-26.09) and the area under the curve was 0.249 (SE = 0.138). The positive and negative predictive values were 5.38 (95% CI 3.89-7.44) and 0.34 (95% CI 0.22-0.54), respectively. For single genes, the specificity reached 0.83~0.96, except for RARβ2, but the sensitivity was relatively low for each gene. Significant heterogeneity across the included studies, the potential publication bias for specificity (p = 0.0231), and the need to validate the clinical utility of ctDNA methylation for monitoring treatment response and predicting outcomes in NSCLC patients represent the main limitations of this study. Conclusions: These results provide evidence of the significant potential of ctDNA methylation as a valuable biomarker for improving the diagnosis of NSCLC, advocating for its integration into clinical practice to enhance patient management.
Keywords: circulating free DNA; early diagnosis; methylation; non-small cell lung cancer.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures








References
Publication types
LinkOut - more resources
Full Text Sources

