Polypiperazine-Based Micelles of Mixed Composition for Gene Delivery
- PMID: 39518308
- PMCID: PMC11548379
- DOI: 10.3390/polym16213100
Polypiperazine-Based Micelles of Mixed Composition for Gene Delivery
Abstract
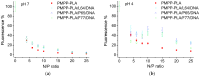
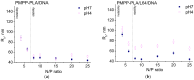
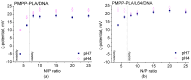
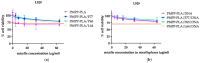
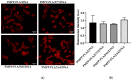
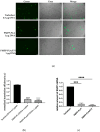
We introduce a novel concept in nucleic acid delivery based on the use of mixed polymeric micelles (MPMs) as platforms for the preparation of micelleplexes with DNA. MPMs were prepared by the co-assembly of a cationic copolymer, poly(1-(4-methylpiperazin-1-yl)-propenone)-b-poly(d,l-lactide), and nonionic poly(ethylene oxide)-b-poly(propylene oxide)-b-poly(ethylene oxide) block copolymers. We hypothesize that by introducing nonionic entities incorporated into the mixed co-assembled structures, the mode and strength of DNA binding and DNA accessibility and release could be modulated. The systems were characterized in terms of size, surface potential, buffering capacity, and binding ability to investigate the influence of composition, in particular, the poly(ethylene oxide) chain length on the properties and structure of the MPMs. Endo-lysosomal conditions were simulated to follow the changes in fundamental parameters and behavior of the micelleplexes. The results were interpreted as reflecting the specific structure and composition of the corona and localization of DNA in the corona, predetermined by the poly(ethylene oxide) chain length. A favorable effect of the introduction of the nonionic block copolymer component in the MPMs and micelleplexes thereof was the enhancement of biocompatibility. The slight reduction of the transfection efficiency of the MPM-based micelleplexes compared to that of the single-component polymer micelles was attributed to the premature release of DNA from the MPM-based micelleplexes in the endo-lysosomal compartments.
Keywords: PEO-PPO-PEO copolymers; cationic copolymers; gene/DNA delivery; internalization/transfection; micelleplexes; mixed polymeric micelles; polyplexes.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

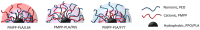




References
-
- Ratner M. First gene therapy approved. Nat. Biotechnol. 2012;30:1153
-
- Ma C.C., Wang Z.L., Xu T., He Z.Y., Wei Y.Q. The approved gene therapy drugs worldwide: From 1998 to 2019. Biotechnol. Adv. 2020;40:107502. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

