Detection of Subarachnoid Hemorrhage Using CNN with Dynamic Factor and Wandering Strategy-Based Feature Selection
- PMID: 39518384
- PMCID: PMC11545384
- DOI: 10.3390/diagnostics14212417
Detection of Subarachnoid Hemorrhage Using CNN with Dynamic Factor and Wandering Strategy-Based Feature Selection
Abstract
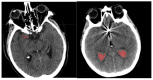
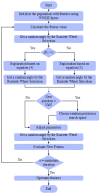
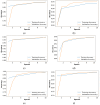
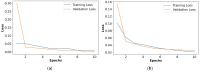
Objectives: Subarachnoid Hemorrhage (SAH) is a serious neurological emergency case with a higher mortality rate. An automatic SAH detection is needed to expedite and improve identification, aiding timely and efficient treatment pathways. The existence of noisy and dissimilar anatomical structures in NCCT images, limited availability of labeled SAH data, and ineffective training causes the issues of irrelevant features, overfitting, and vanishing gradient issues that make SAH detection a challenging task. Methods: In this work, the water waves dynamic factor and wandering strategy-based Sand Cat Swarm Optimization, namely DWSCSO, are proposed to ensure optimum feature selection while a Parametric Rectified Linear Unit with a Stacked Convolutional Neural Network, referred to as PRSCNN, is developed for classifying grades of SAH. The DWSCSO and PRSCNN surpass current practices in SAH detection by improving feature selection and classification accuracy. DWSCSO is proposed to ensure optimum feature selection, avoiding local optima issues with higher exploration capacity and avoiding the issue of overfitting in classification. Firstly, in this work, a modified region-growing method was employed on the patient Non-Contrast Computed Tomography (NCCT) images to segment the regions affected by SAH. From the segmented regions, the wide range of patterns and irregularities, fine-grained textures and details, and complex and abstract features were extracted from pre-trained models like GoogleNet, Visual Geometry Group (VGG)-16, and ResNet50. Next, the PRSCNN was developed for classifying grades of SAH which helped to avoid the vanishing gradient issue. Results: The DWSCSO-PRSCNN obtained a maximum accuracy of 99.48%, which is significant compared with other models. The DWSCSO-PRSCNN provides an improved accuracy of 99.62% in CT dataset compared with the DL-ICH and GoogLeNet + (GLCM and LBP), ResNet-50 + (GLCM and LBP), and AlexNet + (GLCM and LBP), which confirms that DWSCSO-PRSCNN effectively reduces false positives and false negatives. Conclusions: the complexity of DWSCSO-PRSCNN was acceptable in this research, for while simpler approaches appeared preferable, they failed to address problems like overfitting and vanishing gradients. Accordingly, the DWSCSO for optimized feature selection and PRSCNN for robust classification were essential for handling these challenges and enhancing the detection in different clinical settings.
Keywords: feature selection; parametric rectified linear unit; region-growing method; sand cat swarm optimization algorithm; stacked convolutional neural network; subarachnoid hemorrhage detection; wandering strategy; water waves dynamic factor.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
-
- Stetzuhn M., Tigges T., Pielmus A.G., Spies C., Middel C., Klum M., Zaunseder S., Orglmeister R., Feldheiser A. Detection of a Stroke Volume Decrease by Machine-Learning Algorithms Based on Thoracic Bioimpedance in Experimental Hypovolaemia. Sensors. 2022;22:5066. doi: 10.3390/s22145066. - DOI - PMC - PubMed
-
- Kaur S., Singh A. A New Deep Learning Framework for Accurate Intracranial Brain Hemorrhage Detection and Classification Using Real-Time Collected NCCT Images. Appl. Magn. Reson. 2024;55:629–661. doi: 10.1007/s00723-024-01661-z. - DOI
-
- Yu D., Williams G.W., Aguilar D., Yamal J., Maroufy V., Wang X., Zhang C., Huang Y., Gu Y., Talebi Y., et al. Machine Learning Prediction of the Adverse Outcome for Nontraumatic Subarachnoid Hemorrhage Patients. Ann. Clin. Transl. Neurol. 2020;7:2178–2185. doi: 10.1002/acn3.51208. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

