Molecular, Histological, and Functional Changes in Acta1-MCM;FLExDUX4/+ Mice
- PMID: 39518930
- PMCID: PMC11545788
- DOI: 10.3390/ijms252111377
Molecular, Histological, and Functional Changes in Acta1-MCM;FLExDUX4/+ Mice
Abstract
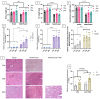
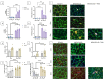
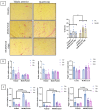
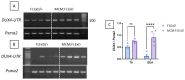
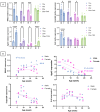
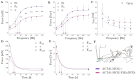
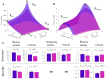
DUX4 is the major gene responsible for facioscapulohumeral dystrophy (FSHD). Several mouse models expressing DUX4 have been developed, the most commonly used by academic laboratories being ACTA1-MCM/FLExDUX4. In this study, molecular and histological modifications in the tibialis anterior and quadriceps muscles were investigated in this model at different time points. We investigated several changes that could be used as markers of therapeutic efficacy. Our results confirm the progressive muscular dystrophy previously described but also highlight biases associated with tamoxifen injections and the complexity of choosing the genes used to calculate a DUX4-pathway gene composite score. We also developed a comprehensive force test that better reflects the movements made in everyday life. This functional force-velocity-endurance model, which describes the force production capacities at all velocity and fatigue levels, was applied on 12-13-week-old animals without tamoxifen. Our data highlight that previously unsuspected muscle properties are also affected by the expression of DUX4, leading to a weaker muscle with a lower initial muscle force but with preserved power and endurance capacity. Importantly, this force-velocity-endurance approach can be used in humans for clinical evaluations.
Keywords: DUX4; FSHD; biomarker; force test; mouse model; muscle; myopathy; strength.
Conflict of interest statement
The authors declare no competing interests.
Figures
References
-
- Orphanet Prevalence and Incidence of Rare Diseases: Bibliographic Data. 2024. [(accessed on 15 October 2024)]. Available online: https://www.orpha.net/pdfs/orphacom/cahiers/docs/GB/Prevalence_of_rare_d....
-
- Gabriels J., Beckers M.C., Ding H., De Vriese A., Plaisance S., van der Maarel S.M., Padberg G.W., Frants R.R., Hewitt J.E., Collen D., et al. Nucleotide sequence of the partially deleted D4Z4 locus in a patient with FSHD identifies a putative gene within each 3.3 kb element. Gene. 1999;236:25–32. doi: 10.1016/S0378-1119(99)00267-X. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

