Integrative Metabolome and Proteome Analysis of Cerebrospinal Fluid in Parkinson's Disease
- PMID: 39518959
- PMCID: PMC11547079
- DOI: 10.3390/ijms252111406
Integrative Metabolome and Proteome Analysis of Cerebrospinal Fluid in Parkinson's Disease
Abstract
Parkinson's disease (PD) is a common neurodegenerative disorder characterized by the loss of dopaminergic neurons in the substantia nigra. Recent studies have highlighted the significant role of cerebrospinal fluid (CSF) in reflecting pathophysiological PD brain conditions by analyzing the components of CSF. Based on the published literature, we created a single network with altered metabolites in the CSF of patients with PD. We analyzed biological functions related to the transmembrane of mitochondria, respiration of mitochondria, neurodegeneration, and PD using a bioinformatics tool. As the proteome reflects phenotypes, we collected proteome data based on published papers, and the biological function of the single network showed similarities with that of the metabolomic network. Then, we analyzed the single network of integrated metabolome and proteome. In silico predictions based on the single network with integrated metabolomics and proteomics showed that neurodegeneration and PD were predicted to be activated. In contrast, mitochondrial transmembrane activity and respiration were predicted to be suppressed in the CSF of patients with PD. This review underscores the importance of integrated omics analyses in deciphering PD's complex biochemical networks underlying neurodegeneration.
Keywords: Parkinson’s disease; cerebrospinal fluid; integrated omics; metabolomics; proteomics.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures
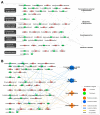

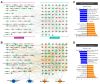
References
-
- Leite Silva A.B.R., Goncalves de Oliveira R.W., Diogenes G.P., de Castro Aguiar M.F., Sallem C.C., Lima M.P.P., de Albuquerque Filho L.B., Peixoto de Medeiros S.D., Penido de Mendonca L.L., de Santiago Filho P.C., et al. Premotor, nonmotor and motor symptoms of Parkinson’s Disease: A new clinical state of the art. Ageing Res. Rev. 2023;84:101834. doi: 10.1016/j.arr.2022.101834. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

