NRXN1 as a Prognostic Biomarker: Linking Copy Number Variation to EMT and Survival in Colon Cancer
- PMID: 39518976
- PMCID: PMC11546699
- DOI: 10.3390/ijms252111423
NRXN1 as a Prognostic Biomarker: Linking Copy Number Variation to EMT and Survival in Colon Cancer
Abstract
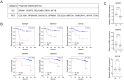
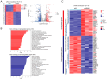
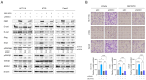
The role of biomarkers in cancer treatment varies significantly depending on the cancer stage. Thus, in clinical practice, tailoring biomarkers to meet the specific needs and challenges of each cancer stage can increase the precision of treatment. Because they reflect underlying genetic alterations that influence cancer progression, copy number variation (CNV) biomarkers can play crucial prognostic roles. In our previous study, we identified potential survival-related genes for colorectal cancer (CRC) by analyzing CNV and gene expression data using a machine-learning approach. To further investigate the biological function of NRXN1, we assessed the use of RNA sequencing, phosphokinase assays, real-time quantitative PCR, and Western blot analysis. We found that NRXN1 copy number deletion was significantly associated with poor overall survival (OS) and recurrence-free survival (RFS), even in patients who received adjuvant chemotherapy. Compared with its expression in normal tissues, NRXN1 expression was lower in tumors, suggesting its potential role as a tumor suppressor. NRXN1 knockdown enhanced CRC cell viability and invasion, and transcriptome analysis indicated that the increased invasion was caused by GSK3β-mediated epithelial-mesenchymal transition. These findings highlight NRXN1 copy number deletion as a novel biomarker for predicting recurrence and survival in patients with resected colon cancer.
Keywords: colorectal cancer (CRC); epithelial–mesenchymal transition; mechanisms of inhibition.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




Similar articles
-
Investigation of ENO2 as a promising novel marker for the progression of colorectal cancer with microsatellite instability-high.BMC Cancer. 2024 May 9;24(1):573. doi: 10.1186/s12885-024-12332-4. BMC Cancer. 2024. PMID: 38724951 Free PMC article.
-
[Prenatal diagnosis of partial deletion of NRXN1 gene with combined CNV-seq and qPCR assays].Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2022 Nov 10;39(11):1200-1204. doi: 10.3760/cma.j.cn511374-20211109-00892. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2022. PMID: 36317203 Chinese.
-
SERPINC1, a new prognostic predictor of colon cancer, promote colon cancer progression through EMT.Cancer Rep (Hoboken). 2024 Jun;7(6):e2079. doi: 10.1002/cnr2.2079. Cancer Rep (Hoboken). 2024. PMID: 38923313 Free PMC article.
-
Colon cancer at the molecular level--usefulness of epithelial-mesenchymal transition analysis.Rev Med Chir Soc Med Nat Iasi. 2012 Oct-Dec;116(4):1106-11. Rev Med Chir Soc Med Nat Iasi. 2012. PMID: 23700897 Review.
-
Novel biomarkers for patient stratification in colorectal cancer: A review of definitions, emerging concepts, and data.World J Gastrointest Oncol. 2018 Jul 15;10(7):145-158. doi: 10.4251/wjgo.v10.i7.145. World J Gastrointest Oncol. 2018. PMID: 30079141 Free PMC article. Review.
References
-
- André T., Boni C., Navarro M., Tabernero J., Hickish T., Topham C., Bonetti A., Clingan P., Bridgewater J., Rivera F., et al. Improved overall survival with oxaliplatin, fluorouracil, and leucovorin as adjuvant treatment in stage II or III colon cancer in the MOSAIC trial. J. Clin. Oncol. 2009;27:3109–3116. doi: 10.1200/JCO.2008.20.6771. - DOI - PubMed
-
- Kotani D., Oki E., Nakamura Y., Yukami H., Mishima S., Bando H., Shirasu H., Yamazaki K., Watanabe J., Kotaka M., et al. Molecular residual disease and efficacy of adjuvant chemotherapy in patients with colorectal cancer. Nat. Med. 2023;29:127–134. doi: 10.1038/s41591-022-02115-4. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

