Pan-Genome-Wide Investigation and Co-Expression Network Analysis of HSP20 Gene Family in Maize
- PMID: 39519102
- PMCID: PMC11546149
- DOI: 10.3390/ijms252111550
Pan-Genome-Wide Investigation and Co-Expression Network Analysis of HSP20 Gene Family in Maize
Abstract
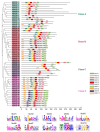
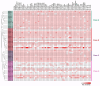
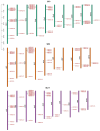
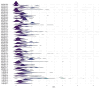
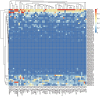
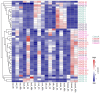
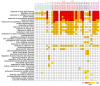
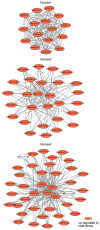
Heat shock protein 20 (HSP20) is a diverse and functionally important protein family that plays a crucial role in plants' tolerance to various abiotic stresses. In this study, we systematically analyzed the structural and functional characteristics of the HSP20 gene family within the Zea pan-genome. By identifying 56 HSP20 pan-genes, we revealed the variation in the number of these genes across different maize inbreds or relatives. Among those 56 genes, only 31 are present in more than 52 inbreds or relatives. Further phylogenetic analysis classified these genes into four major groups (Class A, B, C, D) and explored their diversity in subcellular localization, physicochemical properties, and the terminal structures of those HSP20s. Through collinearity analysis and Ka/Ks ratio calculations, we found that most HSP20 genes underwent purifying selection during maize domestication, although a few genes showed signs of positive selection pressure. Additionally, expression analysis showed that several HSP20 genes were significantly upregulated under high temperatures, particularly in tassels and leaves. Co-expression network analysis revealed that HSP20 genes were significantly enriched in GO terms related to environmental stress responses, suggesting that HSP20 genes not only play key roles in heat stress but may also be involved in regulating various other biological processes, such as secondary metabolism and developmental processes. These findings expand our understanding of the functions of the maize HSP20 family and provide new insights for further research into maize's response mechanisms to environmental stresses.
Keywords: HSP20; Zea pan-genome; co-expression; gene family; heat stress.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
ZmHsp18 screened from the ZmHsp20 gene family confers thermotolerance in maize.BMC Plant Biol. 2024 Nov 6;24(1):1048. doi: 10.1186/s12870-024-05763-5. BMC Plant Biol. 2024. PMID: 39506700 Free PMC article.
-
Phylogenetic and expression analysis of HSP20 gene family in Rhododendron species of different altitudes.Int J Biol Macromol. 2025 May;309(Pt 4):143125. doi: 10.1016/j.ijbiomac.2025.143125. Epub 2025 Apr 12. Int J Biol Macromol. 2025. PMID: 40228764
-
Comprehensive characterization of poplar HSP20 gene family: genome-wide identification, stress-induced expression profiling, and protein interaction verifications.BMC Plant Biol. 2025 Feb 25;25(1):251. doi: 10.1186/s12870-025-06264-9. BMC Plant Biol. 2025. PMID: 39994524 Free PMC article.
-
Genome-wide identification and analysis of biotic and abiotic stress regulation of small heat shock protein (HSP20) family genes in bread wheat.J Plant Physiol. 2017 Apr;211:100-113. doi: 10.1016/j.jplph.2017.01.004. Epub 2017 Jan 22. J Plant Physiol. 2017. PMID: 28178571
-
Genome-wide analysis of the potato Hsp20 gene family: identification, genomic organization and expression profiles in response to heat stress.BMC Genomics. 2018 Jan 18;19(1):61. doi: 10.1186/s12864-018-4443-1. BMC Genomics. 2018. PMID: 29347912 Free PMC article.
Cited by
-
Identification and Bioinformatics Analysis of the HSP20 Family in the Peony.Genes (Basel). 2025 Jun 26;16(7):742. doi: 10.3390/genes16070742. Genes (Basel). 2025. PMID: 40725398 Free PMC article.
References
-
- Kotak S., Port M., Ganguli A., Bicker F., Von Koskull-Döring P. Characterization of C-Terminal Domains of Arabidopsis Heat Stress Transcription Factors (Hsfs) and Identification of a New Signature Combination of Plant Class A Hsfs with AHA and NES Motifs Essential for Activator Function and Intracellular Localization. Plant J. 2004;39:98–112. doi: 10.1111/j.1365-313X.2004.02111.x. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

