Identification of Genes Associated with Familial Focal Segmental Glomerulosclerosis Through Transcriptomics and In Silico Analysis, Including RPL27, TUBB6, and PFDN5
- PMID: 39519211
- PMCID: PMC11546068
- DOI: 10.3390/ijms252111659
Identification of Genes Associated with Familial Focal Segmental Glomerulosclerosis Through Transcriptomics and In Silico Analysis, Including RPL27, TUBB6, and PFDN5
Abstract
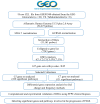
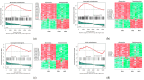
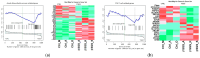
Focal segmental glomerulosclerosis (FSGS) is a major cause of nephrotic syndrome and often leads to progressive kidney failure. Its varying clinical presentation suggests potential genetic diversity, requiring further molecular investigation. This study aims to elucidate some of the genetic and molecular mechanisms underlying FSGS. The study focuses on the use of bioinformatic analysis of gene expression data to identify genes associated with familial FSGS. A comprehensive in silico analysis was performed using the GSE99340 data set from Gene Expression Omnibus (GEO) comparing gene expression in glomerular and tubulointerstitial tissues from FSGS patients (n = 10) and Minimal Change Disease (MCD) patients (n = 8). These findings were validated using transcriptomics data obtained using RNA sequencing from FSGS (n = 3) and control samples (n = 3) from the UAE. Further validation was conducted using qRT-PCR on an independent FFPE cohort (FSGS, n = 6; MCD, n = 7) and saliva samples (FSGS, n = 3; Control, n = 7) from the UAE. Three genes (TUBB6, RPL27, and PFDN5) showed significant differential expression (p < 0.01) when comparing FSGS and MCD with healthy controls. These genes are associated with cell junction organization and synaptic pathways of the neuron, supporting the link between FSGS and the neural system. These genes can potentially be useful as diagnostic biomarkers for FSGS and to develop new treatment options.
Keywords: Focal segmental glomerulosclerosis (FSGS); Minimal change disease (MCD); genetic; glomerular; tubulointerstitial.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


Similar articles
-
Renal tubular gen e biomarkers identification based on immune infiltrates in focal segmental glomerulosclerosis.Ren Fail. 2022 Dec;44(1):966-986. doi: 10.1080/0886022X.2022.2081579. Ren Fail. 2022. PMID: 35713363 Free PMC article.
-
Circulating and urinary microRNA profile in focal segmental glomerulosclerosis: a pilot study.Eur J Clin Invest. 2015 Apr;45(4):394-404. doi: 10.1111/eci.12420. Eur J Clin Invest. 2015. PMID: 25682967 Free PMC article.
-
Transcriptome meta-analysis and validation to discovery of hub genes and pathways in focal and segmental glomerulosclerosis.BMC Nephrol. 2024 Sep 4;25(1):293. doi: 10.1186/s12882-024-03734-4. BMC Nephrol. 2024. PMID: 39232654 Free PMC article.
-
Hidden genetics behind glomerular scars: an opportunity to understand the heterogeneity of focal segmental glomerulosclerosis?Pediatr Nephrol. 2024 Jun;39(6):1685-1707. doi: 10.1007/s00467-023-06046-1. Epub 2023 Sep 20. Pediatr Nephrol. 2024. PMID: 37728640 Free PMC article. Review.
-
Rituximab therapy for focal segmental glomerulosclerosis and minimal change disease in adults: a systematic review and meta-analysis.BMC Nephrol. 2020 Apr 15;21(1):134. doi: 10.1186/s12882-020-01797-7. BMC Nephrol. 2020. PMID: 32293308 Free PMC article.
References
-
- Kalantar-Zadeh K., Baker C.L., Copley J.B., Levy D.I., Berasi S., Tamimi N., Alvir J., Udani S.M. A Retrospective Study of Clinical and Economic Burden of Focal Segmental Glomerulosclerosis (FSGS) in the United States. Kidney Int. Rep. 2021;6:2679. doi: 10.1016/j.ekir.2021.07.030. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

