Concordance Between Biochemical and Molecular Diagnosis Obtained by WES in Mexican Patients with Inborn Errors of Intermediary Metabolism: Utility for Therapeutic Management
- PMID: 39519275
- PMCID: PMC11546494
- DOI: 10.3390/ijms252111722
Concordance Between Biochemical and Molecular Diagnosis Obtained by WES in Mexican Patients with Inborn Errors of Intermediary Metabolism: Utility for Therapeutic Management
Abstract
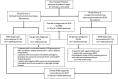
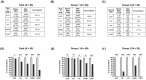
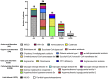
Biochemical phenotyping has been the milestone for diagnosing and managing patients affected by inborn errors of intermediary metabolism (IEiM); however, identifying the genotype responsible for these monogenic disorders greatly contributes to achieving these goals. Herein, whole-exome sequencing (WES) was used to determine the genotypes of 95 unrelated Mexican pediatric patients suspected of having IEiM. They were classified into those bearing specific biochemical abnormalities (Group 1), and those presenting unspecific biochemical profiles (Group 2). The overall concordance between the initial biochemical diagnosis and final genotypic diagnoses was 72.6% (N = 69/95 patients), with the highest concordance achieved in Group 1 (91.3%, N = 63/69), whereas the concordance was limited in Group 2 (23.07%). This finding suggests that previous biochemical phenotyping correlated with the high WES diagnostic success. Concordance was high for urea cycle disorders (94.1%) and organic acid disorders (77.4%). The identified mutational spectrum comprised 83 IEiM-relevant variants (pathogenic, likely pathogenic, and variants of uncertain significance or VUS), including three novel ones, distributed among 29 different genes responsible for amino acid, organic acid, urea cycle, carbohydrate, and lipid disorders. Inconclusive WES results (7.3%, N = 7/95) relied on monoallelic pathogenic genotypes or those involving two VUS for autosomal-recessive IEiMs. A second monogenic disease was observed in 10.5% (N = 10/95) of the patients. According to the WES results, modifications in treatment had to be made in 33.6% (N = 32/95) of patients, mainly attributed to the presence of a second monogenic disease, or to an actionable trait. This study includes the largest cohort of Mexican patients to date with biochemically suspected IEiM who were genetically diagnosed through WES, underscoring its importance in medical management.
Keywords: diagnostic odyssey; genomic medicine; inborn errors of metabolism; personalized medicine; precision medicine; rare diseases; whole-exome sequencing.
Conflict of interest statement
Authors Seung Woo Ryu and Hane Lee were employed by the company 3billion, Inc. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The authors declare no conflicts of interest.
Figures
Similar articles
-
Whole exome sequencing diagnosis of inborn errors of metabolism and other disorders in United Arab Emirates.Orphanet J Rare Dis. 2016 Jul 8;11(1):94. doi: 10.1186/s13023-016-0474-3. Orphanet J Rare Dis. 2016. PMID: 27391121 Free PMC article.
-
A comprehensive multiplex PCR based exome-sequencing assay for rapid bloodspot confirmation of inborn errors of metabolism.BMC Med Genet. 2019 Jan 6;20(1):3. doi: 10.1186/s12881-018-0731-5. BMC Med Genet. 2019. PMID: 30612563 Free PMC article.
-
[Characterization of inborn errors of intermediary metabolism in mexican patients].An Pediatr (Barc). 2014 May;80(5):310-6. doi: 10.1016/j.anpedi.2013.09.003. Epub 2013 Oct 17. An Pediatr (Barc). 2014. PMID: 24140120 Spanish.
-
Update on breastfeeding in newborns with inborn errors of intermediary metabolism.Bol Med Hosp Infant Mex. 2022;79(3):141-151. doi: 10.24875/BMHIM.21000103. Bol Med Hosp Infant Mex. 2022. PMID: 35882023 Review. English.
-
Diagnosis of inborn errors of metabolism through prenatal exome sequencing with targeted analysis for fetal structural anomalies.Prenat Diagn. 2024 Apr;44(4):432-442. doi: 10.1002/pd.6476. Epub 2023 Dec 8. Prenat Diagn. 2024. PMID: 38063435 Review.
References
-
- Wojcik M.H., Reuter C.M., Marwaha S., Mahmoud M., Duyzend M.H., Barseghyan H., Yuan B., Boone P.M., Groopman E.E., Délot E.C., et al. Beyond the exome: What’s next in diagnostic testing for Mendelian conditions. Am. J. Hum. Genet. 2023;110:1229–1248. doi: 10.1016/j.ajhg.2023.06.009. - DOI - PMC - PubMed
-
- Suzuki H., Nozaki M., Yoshihashi H., Imagawa K., Kajikawa D., Yamada M., Yamaguchi Y., Morisada N., Eguchi M., Ohashi S., et al. Genome analysis in sick neonates and infants: High-yield phenotypes and contribution of small copy number variations. J. Pediatr. 2022;244:38–48.e31. doi: 10.1016/j.jpeds.2022.01.033. - DOI - PubMed
-
- Migliavacca M.P., Sobreira J., Bermeo D., Gomes M., Alencar D., Sussuchi L., Souza C.A., Silva J.S., Kroll J.E., Burger M., et al. Whole genome sequencing as a first-tier diagnostic test for infants in neonatal intensive care units: A pilot study in Brazil. Am. J. Med. Genet. Part A. 2024;194:e63544. doi: 10.1002/ajmg.a.63544. - DOI - PubMed
-
- Farwell K.D., Shahmirzadi L., El-Khechen D., Powis Z., Chao E.C., Davis B.T., Baxter R.M., Zeng W., Mroske C., Parra M.C., et al. Enhanced utility of family-centered diagnostic exome sequencing with inheritance model–based analysis: Results from 500 unselected families with undiagnosed genetic conditions. Genet. Med. 2015;17:578–586. doi: 10.1038/gim.2014.154. - DOI - PubMed
-
- Ediae G.U., Lemire G., Chisholm C., Hartley T., Eaton A., Osmond M., Rojas S.K., Huang L., Gillespie M., Care4Rare Canada Consortium et al. The implementation of an enhanced clinical model to improve the diagnostic yield of exome sequencing for patients with a rare genetic disease: A Canadian experience. Am. J. Med. Genet. Part A. 2023;191:338–347. doi: 10.1002/ajmg.a.63022. - DOI - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

