DeepDualEnhancer: A Dual-Feature Input DNABert Based Deep Learning Method for Enhancer Recognition
- PMID: 39519295
- PMCID: PMC11546905
- DOI: 10.3390/ijms252111744
DeepDualEnhancer: A Dual-Feature Input DNABert Based Deep Learning Method for Enhancer Recognition
Abstract
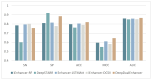
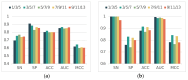
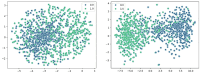
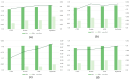
Enhancers are cis-regulatory DNA sequences that are widely distributed throughout the genome. They can precisely regulate the expression of target genes. Since the features of enhancer segments are difficult to detect, we propose DeepDualEnhancer, a DNABert-based method using a multi-scale convolutional neural network, BiLSTM, for enhancer identification. We first designed the DeepDualEnhancer method based only on the DNA sequence input. It mainly consists of a multi-scale Convolutional Neural Network, and BiLSTM to extract features by DNABert and embedding, respectively. Meanwhile, we collected new datasets from the enhancer-promoter interaction field and designed the method DeepDualEnhancer-genomic for inputting DNA sequences and genomic signals, which consists of the transformer sequence attention. Extensive comparisons of our method with 20 other excellent methods through 5-fold cross validation, ablation experiments, and an independent test demonstrated that DeepDualEnhancer achieves the best performance. It is also found that the inclusion of genomic signals helps the enhancer recognition task to be performed better.
Keywords: DNABert; deep learning; enhancer; genomic signal.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures









Similar articles
-
Utilizing a deep learning model based on BERT for identifying enhancers and their strength.PLoS One. 2025 Apr 9;20(4):e0320085. doi: 10.1371/journal.pone.0320085. eCollection 2025. PLoS One. 2025. PMID: 40203028 Free PMC article.
-
iEnhancer-GAN: A Deep Learning Framework in Combination with Word Embedding and Sequence Generative Adversarial Net to Identify Enhancers and Their Strength.Int J Mol Sci. 2021 Mar 30;22(7):3589. doi: 10.3390/ijms22073589. Int J Mol Sci. 2021. PMID: 33808317 Free PMC article.
-
Opening up the blackbox: an interpretable deep neural network-based classifier for cell-type specific enhancer predictions.BMC Syst Biol. 2016 Aug 1;10 Suppl 2(Suppl 2):54. doi: 10.1186/s12918-016-0302-3. BMC Syst Biol. 2016. PMID: 27490187 Free PMC article.
-
Sequence-Based Deep Learning Frameworks on Enhancer-Promoter Interactions Prediction.Curr Pharm Des. 2021;27(15):1847-1855. doi: 10.2174/1381612826666201124112710. Curr Pharm Des. 2021. PMID: 33234095 Review.
-
Application of deep learning in genomics.Sci China Life Sci. 2020 Dec;63(12):1860-1878. doi: 10.1007/s11427-020-1804-5. Epub 2020 Oct 10. Sci China Life Sci. 2020. PMID: 33051704 Review.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

