Isolate-anchored comparisons reveal evolutionary and functional differentiation across SAR86 marine bacteria
- PMID: 39520498
- PMCID: PMC11582366
- DOI: 10.1093/ismejo/wrae227
Isolate-anchored comparisons reveal evolutionary and functional differentiation across SAR86 marine bacteria
Abstract
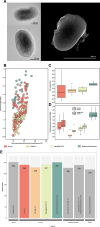
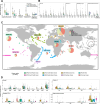
SAR86 is one of the most abundant groups of bacteria in the global surface ocean. However, since its discovery over 30 years ago, it has remained recalcitrant to isolation and many details regarding this group are still unknown. Here, we report the cellular characteristics from the first SAR86 isolate brought into culture, Magnimaribacter mokuoloeensis strain HIMB1674, and use its closed genome in concert with over 700 environmental genomes to assess the phylogenomic and functional characteristics of this order-level lineage of marine Gammaproteobacteria. The SAR86 order Magnimaribacterales invests significant genomic resources into the capacity for $\beta$-oxidation, which is present in most genomes with high gene copy numbers. This cyclical set of reactions appears to be fed by components of cell membranes that include lipids such as phosphatidylcholine, phosphatidylethanolamine, glycolipids, and sulfolipids. In addition to the widespread capacity to degrade the side chain of steroidal compounds via $\beta$-oxidation, several SAR86 sublineages also appear able to fully degrade the steroid polycyclic ring structure as well as other aromatic, polycyclic, and heterocyclic molecules. Read recruitment from publicly available metagenomes reveals that the Magnimaribacterales compose up to 6% of the global surface ocean microbial community. Only a subset of genera drives these high relative abundances, with some more globally dominant and others restricted to specific oceanic regions. This study provides an unprecedented foundation through which to understand this highly abundant yet poorly understood lineage of marine bacteria and charts a path to bring more representatives of this order into laboratory culture.
Keywords: Magnimaribacter; SAR86; marine bacteria; phylogenomics; proteorhodopsin.
© The Author(s) 2024. Published by Oxford University Press.
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Genomic insights to SAR86, an abundant and uncultivated marine bacterial lineage.ISME J. 2012 Jun;6(6):1186-99. doi: 10.1038/ismej.2011.189. Epub 2011 Dec 15. ISME J. 2012. PMID: 22170421 Free PMC article.
-
Global ecotypes in the ubiquitous marine clade SAR86.ISME J. 2020 Jan;14(1):178-188. doi: 10.1038/s41396-019-0516-7. Epub 2019 Oct 14. ISME J. 2020. PMID: 31611653 Free PMC article.
-
Taxonomy and evolution of bacteriochlorophyll a-containing members of the OM60/NOR5 clade of marine gammaproteobacteria: description of Luminiphilus syltensis gen. nov., sp. nov., reclassification of Haliea rubra as Pseudohaliea rubra gen. nov., comb. nov., and emendation of Chromatocurvus halotolerans.BMC Microbiol. 2013 May 24;13:118. doi: 10.1186/1471-2180-13-118. BMC Microbiol. 2013. PMID: 23705883 Free PMC article.
-
The Minderoo-Monaco Commission on Plastics and Human Health.Ann Glob Health. 2023 Mar 21;89(1):23. doi: 10.5334/aogh.4056. eCollection 2023. Ann Glob Health. 2023. PMID: 36969097 Free PMC article. Review.
-
Aequoribacter fuscus gen. nov., sp. nov., a new member of the family Halieaceae, isolated from coastal seawater.J Microbiol. 2020 Jun;58(6):463-471. doi: 10.1007/s12275-020-0206-1. Epub 2020 May 27. J Microbiol. 2020. PMID: 32462487 Review.
References
-
- Mullins TD, Britschgi TB, Krest RLet al. . Genetic comparisons reveal the same unknown bacterial lineages in Atlantic and Pacific bacterioplankton communities. Limnol Oceanogr 1995;40:148–58. 10.4319/lo.1995.40.1.0148 - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

