Identification of metabolic and protein markers representative of the impact of mild nitrogen deficit on agronomic performance of maize hybrids
- PMID: 39520587
- PMCID: PMC11550246
- DOI: 10.1007/s11306-024-02186-z
Identification of metabolic and protein markers representative of the impact of mild nitrogen deficit on agronomic performance of maize hybrids
Abstract
Introduction: A better understanding of the physiological response of silage maize to a mild reduction in nitrogen (N) fertilization and the identification of predictive biochemical markers of N utilization efficiency could contribute to limit the detrimental effect of the overuse of N inputs.
Objectives: We integrated phenotypic and biochemical data to interpret the physiology of maize in response to a mild reduction in N fertilization under agronomic conditions and identify predictive leaf metabolic and proteic markers that could be used to pilot and rationalize N fertilization.
Methods: Eco-physiological, developmental and yield-related traits were measured and complemented with metabolomic and proteomic approaches performed on young leaves of a core panel of 29 European genetically diverse dent hybrids cultivated in the field under non-limiting and reduced N fertilization conditions.
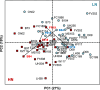
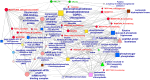
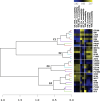
Results: Metabolome and proteome data were analyzed either individually or in an integrated manner together with eco-physiological, developmental, phenotypic and yield-related traits. They allowed to identify (i) common N-responsive metabolites and proteins that could be used as predictive markers to monitor N fertilization, (ii) silage maize hybrids that exhibit improved agronomic performance when N fertilization is reduced.
Conclusions: Among the N-responsive metabolites and proteins identified, a cytosolic NADP-dependent malic enzyme and four metabolite signatures stand out as promising markers that could be used for both breeding and agronomic purposes.
Keywords: Maize; Markers; Metabolome; Nitrogen nutrition; Proteome; Silage.
© 2024. The Author(s).
Conflict of interest statement
The authors have no competing interests to declare that are relevant to the content of this article.
Figures


References
-
- Adegbeye, M. J., Ravi Kanth Reddy, P., Obaisi, A. I., Elghandour, M. M. M. Y., Oyebamiji, K. J., Salem, A. Z. M., Morakinyo-Fasipe, O. T., Cipriano-Salazar, M., & Camacho-Díaz, L. M. (2020). Sustainable agriculture options for production, greenhouse gasses and pollution alleviation, and nutrient recycling in emerging and transitional nations—An overview. Journal of Cleaner Production,242, 118319. 10.1016/j.jclepro.2019.118319 - DOI
-
- Akhi, M. Z., Haque, M. M., & Biswas, M. S. (2021). Role of secondary metabolites to attenuate stress damages in plants. In W. Viduranga (Ed.), Antioxidants (p. 27). IntechOpen.
-
- Amiour, N., Décousset, L., Rouster, J., Quenard, N., Buet, C., Dubreuil, P., Quilleré, I., Brulé, L., Cukier, C., Dinant, S., Sallaud, C., Dubois, F., Limami, A. M., Lea, P. J., & Hirel, B. (2021). Impacts of environmental conditions, and allelic variation of cytosolic glutamine synthetase on maize hybrid kernel production. Communications Biology,4(1), 1095. 10.1038/s42003-021-02598-w - DOI - PMC - PubMed
-
- Amiour, N., Imbaud, S., Clément, G., Agier, N., Zivy, M., Valot, B., Balliau, T., Armengaud, P., Quilleré, I., Cañas, R., Tercet-Laforgue, T., & Hirel, B. (2012). The use of metabolomics integrated with transcriptomic and proteomic studies for identifying key steps involved in the control of nitrogen metabolism in crops such as maize. Journal of Experimental Botany,63(14), 5017–5033. 10.1093/jxb/ers186 - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
