Multiplexed expansion revealing for imaging multiprotein nanostructures in healthy and diseased brain
- PMID: 39521775
- PMCID: PMC11550395
- DOI: 10.1038/s41467-024-53729-w
Multiplexed expansion revealing for imaging multiprotein nanostructures in healthy and diseased brain
Abstract
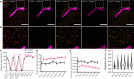
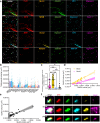
Proteins work together in nanostructures in many physiological contexts and disease states. We recently developed expansion revealing (ExR), which expands proteins away from each other, in order to support better labeling with antibody tags and nanoscale imaging on conventional microscopes. Here, we report multiplexed expansion revealing (multiExR), which enables high-fidelity antibody visualization of >20 proteins in the same specimen, over serial rounds of staining and imaging. Across all datasets examined, multiExR exhibits a median round-to-round registration error of 39 nm, with a median registration error of 25 nm when the most stringent form of the protocol is used. We precisely map 23 proteins in the brain of 5xFAD Alzheimer's model mice, and find reductions in synaptic protein cluster volume, and co-localization of specific AMPA receptor subunits with amyloid-beta nanoclusters. We visualize 20 synaptic proteins in specimens of mouse primary somatosensory cortex. multiExR may be of broad use in analyzing how different kinds of protein are organized amidst normal and pathological processes in biology.
© 2024. The Author(s).
Conflict of interest statement
J.K., M.E.S., and E.S.B. are co-inventors on a patent application for multiExR. The remaining authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
- R01 AG070831/AG/NIA NIH HHS/United States
- R56 AG069192/AG/NIA NIH HHS/United States
- RF1 MH124606/MH/NIMH NIH HHS/United States
- R37 MH080046/MH/NIMH NIH HHS/United States
- 1R01EB024261/U.S. Department of Health & Human Services | NIH | Office of Extramural Research, National Institutes of Health (OER)
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

