Next-generation rapid phenotypic antimicrobial susceptibility testing
- PMID: 39521792
- PMCID: PMC11550857
- DOI: 10.1038/s41467-024-53930-x
Next-generation rapid phenotypic antimicrobial susceptibility testing
Erratum in
-
Publisher Correction: Next-generation rapid phenotypic antimicrobial susceptibility testing.Nat Commun. 2025 Mar 21;16(1):2805. doi: 10.1038/s41467-025-57678-w. Nat Commun. 2025. PMID: 40118838 Free PMC article. No abstract available.
Abstract
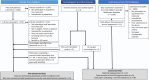
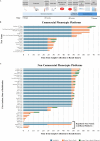
Slow progress towards implementation of conventional clinical bacteriology in low resource settings and strong interest in greater speed for antimicrobial susceptibility testing (AST) more generally has focused attention on next-generation rapid AST technologies. In this Review, we systematically synthesize publications and submissions to regulatory agencies describing technologies that provide phenotypic AST faster than conventional methods. We characterize over ninety technologies in terms of underlying technical innovations, technology readiness level, extent of clinical validation, and time-to-results. This work provides a guide for technology developers and clinical microbiologists to understand the rapid phenotypic AST technology landscape, current development pipeline, and AST-specific validation milestones.
© 2024. The Author(s).
Conflict of interest statement
The authors declare the following competing interests. S.M., D.N., and C.P.Y. co-supervised the work in the publication describing the PhenEXA system. C.P.Y. reports being on an Independent Data Monitoring Committees (IDMC) for Medicago Inc. and InventVacc Biologicals Inc., unrelated to the submitted work. M.P.C. reports personal fees from GEn1E Lifesciences (as a member of the scientific advisory board), personal fees from nplex biosciences (as a member of the scientific advisory board), outside the submitted work. He is the co-founder of Kanvas Biosciences and owns equity in the company. In addition, M.P.C. has a patent Methods for detecting tissue damage, graft versus host disease, and infections using cell-free DNA profiling pending, and a patent Methods for assessing the severity and progression of SARS-CoV-2 infections using cell-free DNA pending. J.P. reports grants from MedImmune, Merck; personal fees from Astra-Zeneca and Merck, all outside the submitted work. All other Authors declare no competing interests.
Figures


References
-
- Petti, C. A., Polage, C. R., Quinn, T. C., Ronald, A. R. & Sande, M. A. Laboratory medicine in Africa: a barrier to effective health care. Clin. Infect. Dis.42, 377–382 (2006). - PubMed
-
- Olmsted, S. S. et al. Strengthening laboratory systems in resource-limited settings. Am. J. Clin. Pathol.134, 374–380 (2010). - PubMed
-
- African Society for Laboratory Medicine (ASLM). Incomplete Antimicrobial Resistance (AMR) Data in Africa: The Crisis Within The Crisis. 12 https://aslm.org/wp-content/uploads/2022/09/ASLM_MAAP-Policy-Brief_Embar... (2022).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

