Novel insights of disulfidptosis-mediated immune microenvironment regulation in atherosclerosis based on bioinformatics analyses
- PMID: 39521794
- PMCID: PMC11550432
- DOI: 10.1038/s41598-024-78392-5
Novel insights of disulfidptosis-mediated immune microenvironment regulation in atherosclerosis based on bioinformatics analyses
Abstract
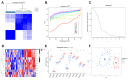
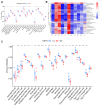
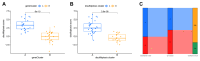
Atherosclerosis (AS) is the leading cause of coronary heart disease, which is the primary cause of death worldwide. Recent studies have identified disulfidptosis as a new type of cell death that may be involved in onset and development of many diseases. However, the role of disulfidptosis in AS is not clear. In this study, bioinformatics analysis and experiments in vivo and in vitro were performed to evaluate the potential relationship between disulfidptosis and AS. AS-related sequencing data were obtained from the Gene Expression Omnibus (GEO). Bioinformatics techniques were used to evaluate differentially expressed genes (DEGs) associated with disulfidptosis-related AS. Hub genes were screened using least absolute shrinkage and selection operator (LASSO) and random forests (RF) methods. In addition, we established a foam cell model in vitro and an AS mouse model in vivo to verify the expressions of hub genes. In addition, we constructed a diagnostic nomogram with hub genes to predict progression of AS. Finally, the consensus clustering method was used to establish two different subtypes, and associations between subtypes and immunity were explored. As the results, 9 disulfidptosis-related AS DEGs were identified from GSE28829 and GSE43292 datasets. Evaluation of DEGs using LASSO and RF methods resulted in identification of 4 hub genes (CAPZB, DSTN, MYL6, PDLIM1), which were analyzed for diagnostic value using ROC curve analysis and verified in vitro and in vivo. Furthermore, a nomogram including hub genes was established that accurately predicted the occurrence of AS. The consensus clustering algorithm was used to separate patients with early atherosclerotic plaques and patients with advanced atherosclerotic plaques into two disulfidptosis subtypes. Cluster B displayed higher levels of infiltrating immune cells, which indicated that patients in cluster B may have a positive immune response for progression of AS. In summary, disulfidptosis-related genes including CAPZB, DSTN, MYL6, and PDLIM1 may be diagnostic markers and therapeutic targets for AS. In addition, these genes are closely related to immune cells, which may inform immunotherapy for AS.
Keywords: Atherosclerosis; Bioinformatics; Consensus cluster; Disulfidptosis; Immune cell infiltration.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

