Establishment of potential lncRNA-related hub genes involved competitive endogenous RNA in lung adenocarcinoma
- PMID: 39522011
- PMCID: PMC11549862
- DOI: 10.1186/s12885-024-13144-2
Establishment of potential lncRNA-related hub genes involved competitive endogenous RNA in lung adenocarcinoma
Abstract
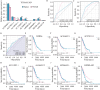
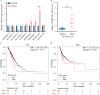
Long non-coding RNAs (lncRNAs) have a notable role in the diagnosis and prognosis of cancer. However, the associations between lncRNA-related hub genes (LRHGs) expression and the corresponding outcomes have not been fully understood in lung adenocarcinoma (LUAD). Here, a total of 71 patients diagnosed with LUAD and 60 healthy volunteers at The First Affiliated Hospital of Huzhou University from April, 2023 to December, 2023 were enrolled in the present study. A LRHGs model was established using least absolute shrinkage and selection operator analyses of The Cancer Genome Atlas-LUAD datasets. The underlying mechanisms of the LRHGs were investigated via Gene Set Enrichment Analysis and Gene Set Variation Analysis. Additionally, the diagnostic role of serum HOXD cluster antisense RNA 2 (HOXD-AS2) was assessed by receiver operating characteristic (ROC) curve analysis. Lastly, TCGA-LUAD samples were divided into high- and low-HOXD-AS2 expression groups based on the median expression. The associations between HOXD-AS2 expression and miR-4538 as well as Calmodulin-Dependent Protein Kinase Type II subunit Beta (CAMK2B) levels were conducted through Pearson correlation analysis. A comprehensive analysis identified 141 differentially expressed lncRNAs between 539 LUAD tissues and 59 normal samples. A prognostic marker for overall survival was established by constructing a predictive signature consisting of 9 LRHGs. Subsequently, 474 LUAD samples were categorized into a high or low-risk group based on the median of the risk score. An independent prognostic model was constructed to confirm the validity of this categorization. Further comparisons of the clinicopathological features and LRHG-related pathways were performed between the two groups. Examinations of LRHG expression in two LUAD clusters and of the association between LRHG expression and immune infiltration were also conducted. HOXD-AS2 expression was shown to be elevated in LUAD tissues compared with matched normal tissues, and the serum HOXD-AS2 level was also notably increased in LUAD samples compared with healthy controls. The results of the ROC analysis indicated that the sensitivity and specificity of HOXD-AS2 were higher than that of cytokeratin-19 fragment (CYFRA21-1), which is a serum marker for LUAD. Pearson analyses indicated that miR-4538 level was negatively associated with HOXD-AS2 expression, but CAMK2B level showed positive correlation in LUAD. The results of the present study therefore indicated that the constructed LRHG model, particularly HOXD-AS2, could independently diagnose and predict the prognosis of LUAD, which suggested the underlying mechanism of the HOXD-AS2/miR-4538/CAMK2B, and might offer efficient strategies for LUAD treatment.
Keywords: Diagnosis; LncRNA; Lung adenocarcinoma; ceRNA.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures










Similar articles
-
Predictions of the dysregulated competing endogenous RNA signature involved in the progression of human lung adenocarcinoma.Cancer Biomark. 2020;29(3):399-416. doi: 10.3233/CBM-200133. Cancer Biomark. 2020. PMID: 32741804
-
A methylation-related lncRNA-based prediction model in lung adenocarcinomas.Clin Respir J. 2024 Aug;18(8):e13753. doi: 10.1111/crj.13753. Clin Respir J. 2024. PMID: 39187946 Free PMC article.
-
USF1-induced overexpression of long noncoding RNA WDFY3-AS2 promotes lung adenocarcinoma progression via targeting miR-491-5p/ZNF703 axis.Mol Carcinog. 2020 Aug;59(8):875-885. doi: 10.1002/mc.23181. Epub 2020 Apr 10. Mol Carcinog. 2020. PMID: 32275336
-
Prognostic risk model of six m7 g-related lncRNAs in lung adenocarcinoma.Eur J Med Res. 2025 Jun 10;30(1):468. doi: 10.1186/s40001-025-02744-8. Eur J Med Res. 2025. PMID: 40490831 Free PMC article.
-
Long noncoding HOXD-AS1: a crucial regulator of malignancy.Front Cell Dev Biol. 2025 Mar 26;13:1543915. doi: 10.3389/fcell.2025.1543915. eCollection 2025. Front Cell Dev Biol. 2025. PMID: 40206400 Free PMC article. Review.
Cited by
-
Development and validation of prognostic models based on cell cycle-related signatures for predicting the prognosis of patients with lung adenocarcinoma.Transl Cancer Res. 2025 May 30;14(5):2900-2915. doi: 10.21037/tcr-24-1479. Epub 2025 May 27. Transl Cancer Res. 2025. PMID: 40530147 Free PMC article.
-
Single-cell RNA sequencing technology was employed to construct a risk prediction model for genes associated with pyroptosis and ferroptosis in lung adenocarcinoma.Respir Res. 2025 Jul 18;26(1):249. doi: 10.1186/s12931-025-03323-5. Respir Res. 2025. PMID: 40682067 Free PMC article.
References
-
- Siegel RL, Miller KD, Wagle NS and Jemal A: Cancer statistics, 2023. CA: a cancer journal for clinicians 73: 17–48, 2023. - PubMed
-
- Sung H, Ferlay J, Siegel RL, et al.: Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA: a cancer journal for clinicians 71: 209–249, 2021. - PubMed
MeSH terms
Substances
Grants and funding
- 2023KY318/The Scientific Technology Projects of Health and Medicine of Zhejiang Province under Grant nos
- 2023KY1180/The Scientific Technology Projects of Health and Medicine of Zhejiang Province under Grant nos
- LTGY23H160004/Zhejiang Provincial Natural Science Foundation of China under Grant Nos
- LY22H160026/Zhejiang Provincial Natural Science Foundation of China under Grant Nos
- 2021GZB03/Huzhou Science and Technology Fund under Grant no
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

