Knockdown Proteomics Reveals USP7 as a Regulator of Cell-Cell Adhesion in Colorectal Cancer via AJUBA
- PMID: 39522755
- PMCID: PMC11697772
- DOI: 10.1016/j.mcpro.2024.100878
Knockdown Proteomics Reveals USP7 as a Regulator of Cell-Cell Adhesion in Colorectal Cancer via AJUBA
Abstract
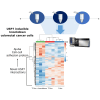
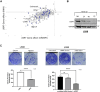
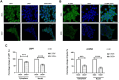
Ubiquitin-specific protease 7 (USP7) is implicated in many cancers including colorectal cancer in which it regulates cellular pathways such as Wnt signaling and the P53-MDM2 pathway. With the discovery of small-molecule inhibitors, USP7 has also become a promising target for cancer therapy and therefore systematically identifying USP7 deubiquitinase interaction partners and substrates has become an important goal. In this study, we selected a colorectal cancer cell model that is highly dependent on USP7 and in which USP7 knockdown significantly inhibited colorectal cancer cell viability, colony formation, and cell-cell adhesion. We then used inducible knockdown of USP7 followed by LC-MS/MS to quantify USP7-dependent proteins. We identified the Ajuba LIM domain protein as an interacting partner of USP7 through co-IP, its substantially reduced protein levels in response to USP7 knockdown, and its sensitivity to the specific USP7 inhibitor FT671. The Ajuba protein has been shown to have oncogenic functions in colorectal and other tumors, including regulation of cell-cell adhesion. We show that both knockdown of USP7 or Ajuba results in a substantial reduction of cell-cell adhesion, with concomitant effects on other proteins associated with adherens junctions. Our findings underlie the role of USP7 in colorectal cancer through its protein interaction networks and show that the Ajuba protein is a component of USP7 protein networks present in colorectal cancer.
Keywords: AJUBA; USP7; cancer; deubiquitinase; inducible knock-down.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflicts of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures






References
-
- Dewson G., Eichhorn P.J.A., Komander D. Deubiquitinases in cancer. Nat. Rev. Cancer. 2023;23:842–862. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

