Replenishing co-downregulated miR-100-5p and miR-125b-5p in malignant germ cell tumors causes growth inhibition through cell cycle disruption
- PMID: 39522951
- PMCID: PMC11977657
- DOI: 10.1002/1878-0261.13757
Replenishing co-downregulated miR-100-5p and miR-125b-5p in malignant germ cell tumors causes growth inhibition through cell cycle disruption
Abstract
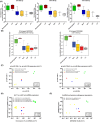
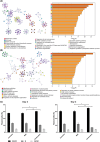
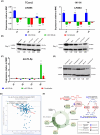
MicroRNAs (miRNAs) are short, nonprotein-coding RNAs, and their expression is dysregulated in malignant germ cell tumors (GCTs). Here, we investigated the causes and consequences of downregulated miR-99a-5p/miR-100-5p (functionally identical) and miR-125b-5p levels in malignant GCTs regardless of age, site, or subtype. Quantitative RT-PCR was used to assess miR-99a-5p/miR-100-5p, miR-125b-5p, and associated gene expression in malignant GCT tissues/cell lines [seminoma (Sem), yolk sac tumor (YST), embryonal carcinoma (EC)]. Cells were treated with demethylating 5-azacytidine and pyrosequencing was performed. Combination miR-100-5p/miR-125b-5p mimic replenishment was used to treat malignant GCT cells. Global messenger RNA (mRNA) targets of the replenished miRNAs were identified and Metascape used to study pathway effects. We found that expression levels of miR-99a-5p/miR-100-5p and miR-125b-5p, their respective pri-miRNAs, and associated genes from chromosomes 11 and 21 (chr11/chr21) were downregulated and highly correlated in malignant GCT cells. Treatment with 5-azacytidine caused upregulation of these miRNAs, with pyrosequencing revealing hypermethylation of their chr11/chr21 loci, likely contributing to miR-100-5p/miR-125b-5p downregulation. Combination miR-100-5p/miR-125b-5p mimic replenishment resulted in growth inhibition in Sem/YST cells, with miR-100-5p/miR-125b-5p mRNA targets enriched in downregulated genes, which were involved in cell cycle (confirmed by flow cytometry) and signaling pathways. Knockdown of the miR-100-5p/miR-125b-5p target tripartite motif containing 71 (TRIM71kd) recapitulated miR-100-5p/miR-125b-5p replenishment, with growth inhibition and cell cycle disruption of Sem/YST/EC cells. Further, replenishment led to reduced lin-28 homolog A (LIN28A) levels and concomitant increases in let-7 (MIRLET7B) tumor suppressor miRNAs, creating a sustained reversion of cell phenotype. In summary, combination miR-100-5p/miR-125b-5p mimic replenishment or TRIM71kd caused growth inhibition in malignant GCT cells via cell cycle disruption. Further studies are now warranted, including mimic treatment alongside conventional platinum-based chemotherapy.
Keywords: germ cell tumor; in vitro models; mRNA; methylation; microRNA; testis.
© 2024 The Author(s). Molecular Oncology published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Teilum G. Classification of endodermal sinus tumour (mesoblatoma vitellinum) and so‐called “embryonal carcinoma” of the ovary. Acta Pathol Microbiol Scand. 1965;64(4):407–429. - PubMed
-
- Coradini PP, Cigana L, Selistre SG, Rosito LS, Brunetto AL. Ototoxicity from cisplatin therapy in childhood cancer. J Pediatr Hematol Oncol. 2007;29(6):355–360. - PubMed
-
- Kollmannsberger C, Kuzcyk M, Mayer F, Hartmann JT, Kanz L, Bokemeyer C. Late toxicity following curative treatment of testicular cancer. Semin Surg Oncol. 1999;17(4):275–281. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

