Nano Plasma Membrane Vesicle-Lipid Nanoparticle Hybrids for Enhanced Gene Delivery and Expression
- PMID: 39523736
- PMCID: PMC11694093
- DOI: 10.1002/adhm.202401888
Nano Plasma Membrane Vesicle-Lipid Nanoparticle Hybrids for Enhanced Gene Delivery and Expression
Abstract
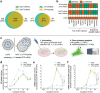
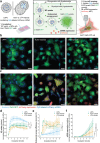
Lipid nanoparticles (LNPs) have emerged as the leading nonviral nucleic acid (NA) delivery system, gaining widespread attention for their use in COVID-19 vaccines. They are recognized for their efficient NA encapsulation, modifiability, and scalable production. However, LNPs face efficacy and potency limitations due to suboptimal intracellular processing, with endosomal escape efficiencies (ESE) below 2.5%. Additionally, up to 70% of NPs undergo recycling and exocytosis after cellular uptake. In contrast, cell-derived vesicles offer biocompatibility and high-delivery efficacy but are challenging to load with exogenous NAs and to manufacture at large-scale. To leverage the strengths of both systems, a hybrid system is designed by combining cell-derived vesicles, such as nano plasma membrane vesicles (nPMVs), with LNPs through microfluidic mixing and subsequent dialysis. These hybrids demonstrate up to tenfold increase in ESE and an 18-fold rise in reporter gene expression in vitro and in vivo in zebrafish larvae (ZFL) and mice, compared to traditional LNPs. These improvements are linked to their unique physico-chemical properties, composition, and morphology. By incorporating cell-derived vesicles, this strategy streamlines the development process, significantly enhancing the efficacy and potency of gene delivery systems without the need for extensive screening.
Keywords: extracellular vesicles; gene delivery; gene therapy; hybrid vesicles; lipid nanoparticles.
© 2024 The Author(s). Advanced Healthcare Materials published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Comirnaty (BioNTech/Pfizer COVID‐19 vaccine) – EMA. https://www.ema.europa.eu/en/documents/assessment-report/comirnaty-epar-..., (accessed: May 2023).
-
- EMA. Spikevax, (previously COVID‐19 Vaccine Moderna). European Medicines Agency https://www.ema.europa.eu/en/medicines/human/EPAR/spikevax 2021. (accessed: April 2023).
-
- Warne N., Ruesch M., Siwik P., Mensah P., Ludwig J., Hripcsak M., Godavarti R., Prigodich A., Dolsten M., Nat. Biotechnol. 2023, 41, 183. - PubMed
-
- Akinc A., Maier M. A., Manoharan M., Fitzgerald K., Jayaraman M., Barros S., Ansell S., Du X., Hope M. J., Madden T. D., Mui B. L., Semple S. C., Tam Y. K., Ciufolini M., Witzigmann D., Kulkarni J. A., van der Meel R., Cullis P. R., Nat. Nanotechnol. 2019, 14, 1084. - PubMed
-
- Arabi F., Mansouri V., Ahmadbeigi N., Biomed. Pharmacother. 2022, 153, 113324. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

