Recent advances in methylation modifications of microRNA
- PMID: 39524539
- PMCID: PMC11550756
- DOI: 10.1016/j.gendis.2023.101201
Recent advances in methylation modifications of microRNA
Abstract
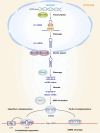
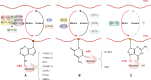
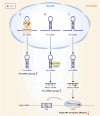
microRNAs (miRNAs) are short single-stranded non-coding RNAs between 21 and 25 nt in length in eukaryotic organisms, which control post-transcriptional gene expression. Through complementary base pairing, miRNAs generally bind to their target messenger RNAs and repress protein production by destabilizing the messenger RNA and translational silencing. They regulate almost all life activities, such as cell proliferation, differentiation, apoptosis, tumorigenesis, and host-pathogen interactions. Methylation modification is the most common RNA modification in eukaryotes. miRNA methylation exists in different types, mainly N6-methyladenosine, 5-methylcytosine, and 7-methylguanine, which can change the expression level and biological mode of action of miRNAs and improve the activity of regulating gene expression in a very fine-tuned way with flexibility. In this review, we will summarize the recent findings concerning methylation modifications of miRNA, focusing on their biogenesis and the potential role of miRNA fate and functions.
Keywords: Methylation modification; m5C modification; m6A modification; m7G modification; miRNA.
© 2023 The Authors. Publishing services by Elsevier B.V. on behalf of KeAi Communications Co., Ltd.
Conflict of interest statement
The authors declared that there was no conflict of interests.
Figures
Similar articles
-
Involvement of RNA methylation modification patterns mediated by m7G, m6A, m5C and m1A regulators in immune microenvironment regulation of Sjögren's syndrome.Cell Signal. 2023 Jun;106:110650. doi: 10.1016/j.cellsig.2023.110650. Epub 2023 Mar 17. Cell Signal. 2023. PMID: 36935085
-
The functions and mechanisms of post-translational modification in protein regulators of RNA methylation: Current status and future perspectives.Int J Biol Macromol. 2023 Dec 31;253(Pt 2):126773. doi: 10.1016/j.ijbiomac.2023.126773. Epub 2023 Sep 9. Int J Biol Macromol. 2023. PMID: 37690652 Review.
-
Role of Main RNA Methylation in Hepatocellular Carcinoma: N6-Methyladenosine, 5-Methylcytosine, and N1-Methyladenosine.Front Cell Dev Biol. 2021 Nov 30;9:767668. doi: 10.3389/fcell.2021.767668. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34917614 Free PMC article. Review.
-
An m6A/m5C/m1A/m7G-Related Long Non-coding RNA Signature to Predict Prognosis and Immune Features of Glioma.Front Genet. 2022 May 26;13:903117. doi: 10.3389/fgene.2022.903117. eCollection 2022. Front Genet. 2022. PMID: 35692827 Free PMC article.
-
The lncRNA epigenetics: The significance of m6A and m5C lncRNA modifications in cancer.Front Oncol. 2023 Mar 9;13:1063636. doi: 10.3389/fonc.2023.1063636. eCollection 2023. Front Oncol. 2023. PMID: 36969033 Free PMC article. Review.
Cited by
-
The roles of LncRNA CARMN in cancers: biomarker potential, therapeutic targeting, and immune response.Discov Oncol. 2024 Dec 18;15(1):776. doi: 10.1007/s12672-024-01679-6. Discov Oncol. 2024. PMID: 39692999 Free PMC article. Review.
-
Serine Hydroxymethyltransferase Modulates Midgut Physiology in Aedes aegypti Through miRNA Regulation: Insights from Small RNA Sequencing and Gene Expression Analysis.Biomolecules. 2025 Apr 30;15(5):644. doi: 10.3390/biom15050644. Biomolecules. 2025. PMID: 40427537 Free PMC article.
-
Emerging Orchestrator of Ecological Adaptation: m6A Regulation of Post-Transcriptional Mechanisms.Mol Ecol. 2025 Aug;34(15):e17545. doi: 10.1111/mec.17545. Epub 2024 Oct 5. Mol Ecol. 2025. PMID: 39367666 Free PMC article. Review.
References
-
- Lee R.C., Feinbaum R.L., Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854. - PubMed
-
- Wightman B., Ha I., Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75(5):855–862. - PubMed
-
- Lee R.C., Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294(5543):862–864. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Research Materials

