Variability of plant transcriptomic responses under stress acclimation: a review from high throughput studies
- PMID: 39524930
- PMCID: PMC11543463
- DOI: 10.3389/abp.2024.13585
Variability of plant transcriptomic responses under stress acclimation: a review from high throughput studies
Abstract
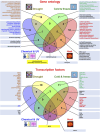
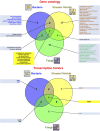
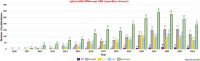
Plant transcriptomes are complex entities shaped spatially and temporally by a multitude of stressors. The aim of this review was to summarize the most relevant transcriptomic responses to selected abiotic (UV radiation, chemical compounds, drought, suboptimal temperature) and biotic (bacteria, fungi, viruses, viroids) stress conditions in a variety of plant species, including model species, crops, and medicinal plants. Selected basic and applicative studies employing RNA-seq from various sequencing platforms and single-cell RNA-seq were involved. The transcriptomic responsiveness of various plant species and the diversity of affected gene families were discussed. Under stress acclimation, plant transcriptomes respond particularly dynamically. Stress response involved both distinct, but also similar gene families, depending on the species, tissue, and the quality and dosage of the stressor. We also noted the over-representation of transcriptomic data for some plant organs. Studies on plant transcriptomes allow for a better understanding of response strategies to environmental conditions. Functional analyses reveal the multitude of stress-affected genes as well as acclimatory mechanisms and suggest metabolome diversity, particularly among medicinal species. Extensive characterization of transcriptomic responses to stress would result in the development of new cultivars that would cope with stress more efficiently. These actions would include modern methodological tools, including advanced genetic engineering, as well as gene editing, especially for the expression of selected stress proteins in planta and for metabolic modifications that allow more efficient synthesis of secondary metabolites.
Keywords: RNA-seq; acclimation; differentially expressed genes; plant transcriptome; stress response.
Copyright © 2024 Rurek and Smolibowski.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Comparative transcriptomic and physiological analyses of contrasting hybrid cultivars ND476 and ZX978 identify important differentially expressed genes and pathways regulating drought stress tolerance in maize.Genes Genomics. 2020 Aug;42(8):937-955. doi: 10.1007/s13258-020-00962-4. Epub 2020 Jul 4. Genes Genomics. 2020. PMID: 32623576
-
Enhancement of Plant Productivity in the Post-Genomics Era.Curr Genomics. 2016 Aug;17(4):295-6. doi: 10.2174/138920291704160607182507. Curr Genomics. 2016. PMID: 27499678 Free PMC article.
-
ESKIMO1 is a key gene involved in water economy as well as cold acclimation and salt tolerance.BMC Plant Biol. 2008 Dec 7;8:125. doi: 10.1186/1471-2229-8-125. BMC Plant Biol. 2008. PMID: 19061521 Free PMC article.
-
Plant hormone-mediated regulation of stress responses.BMC Plant Biol. 2016 Apr 14;16:86. doi: 10.1186/s12870-016-0771-y. BMC Plant Biol. 2016. PMID: 27079791 Free PMC article. Review.
-
Deciphering Macromolecular Interactions Involved in Abiotic Stress Signaling: A Review of Bioinformatics Analysis.Methods Mol Biol. 2023;2642:257-294. doi: 10.1007/978-1-0716-3044-0_15. Methods Mol Biol. 2023. PMID: 36944884 Review.
Cited by
-
Wheat COBRA-like Gene TaCOBL6A2 Confers Heat Tolerance in Plants.Int J Mol Sci. 2025 Apr 25;26(9):4101. doi: 10.3390/ijms26094101. Int J Mol Sci. 2025. PMID: 40362340 Free PMC article.
-
Metabolomics and transcriptomics indicate the changes in medicinal components of Amygdalus mongolica kernels during different developmental stages.Front Plant Sci. 2025 May 23;16:1597638. doi: 10.3389/fpls.2025.1597638. eCollection 2025. Front Plant Sci. 2025. PMID: 40487220 Free PMC article.
-
Diversity of organ-specific plant transcriptomes.Acta Biochim Pol. 2025 Jul 16;72:14609. doi: 10.3389/abp.2025.14609. eCollection 2025. Acta Biochim Pol. 2025. PMID: 40740964 Free PMC article. Review.
-
The Emerging Role of Omics-Based Approaches in Plant Virology.Viruses. 2025 Jul 15;17(7):986. doi: 10.3390/v17070986. Viruses. 2025. PMID: 40733603 Free PMC article. Review.
References
-
- Ascencio-Ibáñez J. T., Sozzani R., Lee T.-J., Chu T.-M., Wolfinger R. D., Cella R., et al. (2008). Global analysis of Arabidopsis gene expression uncovers a complex array of changes impacting pathogen response and cell cycle during geminivirus infection. Plant Physiol. 148, 436–454. 10.1104/pp.108.121038 - DOI - PMC - PubMed
-
- Asseng S., Foster I., Turner N. C. (2011). The impact of temperature variability on wheat yields. Glob. Change Biol. 17, 997–1012. 10.1111/j.1365-2486.2010.02262.x - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

