Single cell and TCR analysis of immune cells from AAV gene therapy-dosed Duchenne muscular dystrophy patients
- PMID: 39524974
- PMCID: PMC11546459
- DOI: 10.1016/j.omtm.2024.101349
Single cell and TCR analysis of immune cells from AAV gene therapy-dosed Duchenne muscular dystrophy patients
Abstract
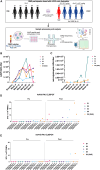
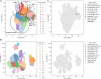
Clinical trials for Duchenne muscular dystrophy (DMD) are assessing the therapeutic efficacy of systemically delivered adeno-associated virus (AAV) carrying a modified DMD transgene. High vector doses (>1E14 vg/kg) are needed to globally transduce skeletal muscles; however, such doses trigger immune-related adverse events. Mitigating these immune responses is crucial for widespread application of AAV-based therapies. We used single-cell RNA sequencing and T cell receptor (TCR) sequencing on peripheral blood mononuclear cells from five participants prior to, and after, dosing. One subject in the high-dose cohort experienced thrombotic microangiopathy (TMA). Few changes in cell frequencies occurred after treatment; however, differential gene expression demonstrated induction of interferon response genes in most T cell types. T cell clonotype and clumping analysis showed the expansion or appearance of groups of related TCR sequences in the post-treatment samples. Three of these expanded clumps could be assigned to prior human herpesvirus infections, two of which were present in the participant that exhibited TMA. These data provide insight on the mechanistic basis of human immune-AAV interactions and lay a foundation for improved understanding of why TMA arises in some patients and not others.
Keywords: AAV; Duchenne; T cell receptor sequencing; adeno-associated virus; gene therapy; immune response; muscular dystrophy; neuromuscular disorders; single-cell sequencing.
© 2024 The Author(s).
Conflict of interest statement
M.J.S. and C.S.Y. are co-founders of MyoGene Bio, a startup spun out of UCLA developing gene editing therapies for DMD. C.S.Y. and M.R.E. are employees of MyoGene Bio. M.A.B. is a listed inventor on a pending patent relating to AAV gene therapy (WO 2021/242664 A1).
Figures




References
-
- Harper S.Q., Hauser M.A., DelloRusso C., Duan D., Crawford R.W., Phelps S.F., Harper H.A., Robinson A.S., Engelhardt J.F., Brooks S.V., Chamberlain J.S. Modular flexibility of dystrophin: implications for gene therapy of Duchenne muscular dystrophy. Nat. Med. 2002;8:253–261. doi: 10.1038/nm0302-253. - DOI - PubMed
-
- Manno C.S., Pierce G.F., Arruda V.R., Glader B., Ragni M., Rasko J.J., Ozelo M.C., Hoots K., Blatt P., Konkle B., et al. Successful transduction of liver in hemophilia by AAV-Factor IX and limitations imposed by the host immune response. Nat. Med. 2006;12:342–347. doi: 10.1038/nm1358. - DOI - PubMed
-
- Salabarria S.M., Corti M., Coleman K.E., Wichman M.B., Berthy J.A., D'Souza P., Tifft C.J., Herzog R.W., Elder M.E., Shoemaker L.R., et al. Thrombotic microangiopathy following systemic AAV administration is dependent on anti-capsid antibodies. J. Clin. Invest. 2024;134 doi: 10.1172/JCI173510. - DOI - PMC - PubMed

