Exploring SSR1 as a novel diagnostic and prognostic biomarker in hepatocellular carcinoma, and its relationship with immune infiltration
- PMID: 39525035
- PMCID: PMC11543030
- DOI: 10.21037/tcr-24-277
Exploring SSR1 as a novel diagnostic and prognostic biomarker in hepatocellular carcinoma, and its relationship with immune infiltration
Abstract
Background: Although signal sequence receptor subunit 1 (SSR1) has undergone thorough examination in different cancer types, its importance in hepatocellular carcinoma (HCC) remains largely uncharted and warrants further investigation. The aim of this study is to explore the role of SSR1 in HCC progression and to decipher its underlying molecular mechanisms.
Methods: We employed the ONCOMINE, Tumor IMmune Estimation Resource (TIMER), and The Cancer Genome Atlas databases to assess SSR1 expression levels within tumor tissues. Logistic and Cox regression analyses, Kaplan-Meier survival plots, nomograms, and forest plots were employed to establish correlation between SSR1 and prognosis. Receiver operating characteristic (ROC) curves demonstrated diagnostic utility of SSR1. Additionally, Gene Ontology (GO) and gene set enrichment analysis (GSEA) analyses were conducted to uncover relevant molecular pathways. TIMER was instrumental in elucidating the connection between SSR1 and immune cell infiltration. Actions of SSR1 in HCC proliferation and migration were investigated through quantitative real-time polymerase chain reaction, Cell Counting Kit-8, 5-ethynyl-2'-deoxyuridine cell proliferation assays, and Transwell migration and wound healing experiments.
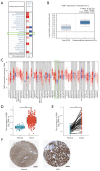
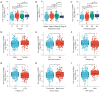
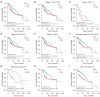
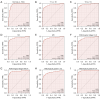
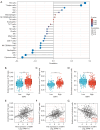
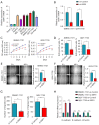
Results: Elevated SSR1 levels were found to be correlated with clinical parameters such as age and pathologic stage, thereby predicting a reduced overall survival (OS) rate in HCC patients. Multivariate survival analysis underscored SSR1 as an independent prognostic marker for OS. A nomogram underscored SSR1's effectiveness as a predictive tool for HCC outcomes, while ROC analysis indicated its high diagnostic accuracy. GO and GSEA analyses suggested that elevated SSR1 expression may be associated with epithelial-mesenchymal transition (EMT) pathway. SSR1 exhibited a negative correlation with cytotoxic cells and a positive correlation with Th2 cells. Our in vitro experiments provided evidence that heightened SSR1 levels may impact HCC proliferation and migration through EMT pathway.
Conclusions: SSR1 surfaces as a new diagnostic and potentially prognostic biomarker, showing an association with immune cell infiltration and cell proliferation in HCC.
Keywords: Signal sequence receptor subunit 1 (SSR1); epithelial-mesenchymal transition (EMT); hepatocellular carcinoma (HCC); immune infiltration.
2024 AME Publishing Company. All rights reserved.
Conflict of interest statement
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-277/coif). The authors have no conflicts of interest to declare.
Figures


References
LinkOut - more resources
Full Text Sources
Miscellaneous
