Determining key residues of engineered scFv antibody variants with improved MMP-9 binding using deep sequencing and machine learning
- PMID: 39525083
- PMCID: PMC11550764
- DOI: 10.1016/j.csbj.2024.10.005
Determining key residues of engineered scFv antibody variants with improved MMP-9 binding using deep sequencing and machine learning
Abstract
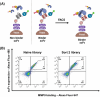
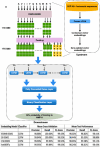
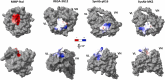
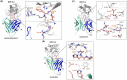
Given the crucial role of specific matrix metalloproteinases (MMPs) in the extracellular matrix, an imbalance in the regulation of activation of matrix metalloproteinase-9 (MMP-9) zymogen and inhibition of the enzyme can result in various diseases, such as cancer, neurodegenerative, and gynecological diseases. Thus, developing novel therapeutics that target MMP-9 with single-chain antibody fragments (scFvs) is a promising approach. We used fluorescent-activated cell sorting (FACS) to screen a synthetic scFv antibody library displayed on yeast for enhanced binding to MMP-9. The screened scFv mutants demonstrated improved binding to MMP-9 compared to the natural inhibitor of MMPs, tissue inhibitor of metalloproteinases (TIMPs). To identify the molecular determinants of these engineered scFv variants that affect binding to MMP-9, we used next-generation DNA sequencing and computational protein structure analysis. Additionally, a deep-learning language model was trained on the screened scFv library of variants to predict the binding affinities of scFv variants based on their CDR-H3 sequences.
Keywords: Antibody engineering; MMP-9; Machine learning; Metalloproteinase; Protein complex structural modeling; Single-chain antibody fragment; Yeast surface display.
© 2024 Published by Elsevier B.V. on behalf of Research Network of Computational and Structural Biotechnology.
Conflict of interest statement
The authors have no conflict of interest.
Figures





Update of
-
Elucidating key determinants of engineered scFv antibody in MMP-9 binding using high throughput screening and machine learning.bioRxiv [Preprint]. 2024 Jun 6:2024.06.04.597476. doi: 10.1101/2024.06.04.597476. bioRxiv. 2024. Update in: Comput Struct Biotechnol J. 2024 Oct 10;23:3759-3770. doi: 10.1016/j.csbj.2024.10.005. PMID: 38895413 Free PMC article. Updated. Preprint.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
