Prediction of conformational states in a coronavirus channel using Alphafold-2 and DeepMSA2: Strengths and limitations
- PMID: 39525089
- PMCID: PMC11543627
- DOI: 10.1016/j.csbj.2024.10.021
Prediction of conformational states in a coronavirus channel using Alphafold-2 and DeepMSA2: Strengths and limitations
Abstract
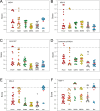
The envelope (E) protein is present in all coronavirus genera. This protein can form pentameric oligomers with ion channel activity which have been proposed as a possible therapeutic target. However, high resolution structures of E channels are limited to those of the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), responsible for the recent COVID-19 pandemic. In the present work, we used Alphafold-2 (AF2), in ColabFold without templates, to predict the transmembrane domain (TMD) structure of six E-channels representative of genera alpha-, beta- and gamma-coronaviruses in the Coronaviridae family. High-confidence models were produced in all cases when combining multiple sequence alignments (MSAs) obtained from DeepMSA2. Overall, AF2 predicted at least two possible orientations of the α-helices in E-TMD channels: one where a conserved polar residue (Asn-15 in the SARS sequence) is oriented towards the center of the channel, 'polar-in', and one where this residue is in an interhelical orientation 'polar-inter'. For the SARS models, the comparison with the two experimental models 'closed' (PDB: 7K3G) and 'open' (PDB: 8SUZ) is described, and suggests a ∼60˚ α-helix rotation mechanism involving either the full TMD or only its N-terminal half, to allow the passage of ions. While the results obtained are not identical to the two high resolution models available, they suggest various conformational states with striking similarities to those models. We believe these results can be further optimized by means of MSA subsampling, and guide future high resolution structural studies in these and other viral channels.
Keywords: AlphaFold-2; Coronavirus; Deep MSA; Envelope protein; IBV; Ion channel; SARS.
© 2024 The Authors.
Conflict of interest statement
The authors have declared no conflict of interest.
Figures






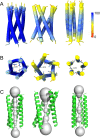
References
-
- Drosten C., Günther S., Preiser W., Van der Werf S., Brodt H.R., Becker S., et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med. 2003;348:1967–1976. - PubMed
-
- Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., et al. A novel coronavirus associated with severe acute respiratory syndrome. New Engl J Med. 2003;348:1953–1966. - PubMed
-
- Zaki A.M., Van Boheemen S., Bestebroer T.M., Osterhaus A.D.M.E., Fouchier R.A.M. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. New Engl J Med. 2012;367:1814–1820. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous
