Ribonucleotide reductase small subunit M2 promotes the proliferation of esophageal squamous cell carcinoma cells via HuR-mediated mRNA stabilization
- PMID: 39525580
- PMCID: PMC11544187
- DOI: 10.1016/j.apsb.2024.07.022
Ribonucleotide reductase small subunit M2 promotes the proliferation of esophageal squamous cell carcinoma cells via HuR-mediated mRNA stabilization
Abstract
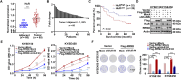
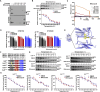
Esophageal squamous cell carcinoma (ESCC), a malignancy of the digestive system, is highly prevalent and the primary cause of cancer-related deaths worldwide due to the lack of early diagnostic biomarkers and effective therapeutic targets. Dysregulated ribonucleotide reductase (RNR) expression has been confirmed to be causally linked to tumorigenesis. This study demonstrated that ribonucleotide reductase small subunit M2 (RRM2) is significantly upregulated in ESCC tissue and that its expression is negatively correlated with clinical outcomes. Mechanistically, HuR promotes RRM2 mRNA stabilization by binding to the adenine/uridine (AU)-rich elements (AREs) within the 3'UTR, resulting in persistent overexpression of RRM2. Furthermore, bifonazole is identified as an inhibitor of HuR via computational screening and molecular docking analysis. Bifonazole disrupts HuR-ARE interactions by competitively binding to HuR at F65, R97, I103, and R153 residues, resulting in reduced RRM2 expression. Furthermore, bifonazole exhibited antitumor effects on ESCC patient-derived xenograft (PDX) models by decreasing RRM2 expression and the dNTP pool. In summary, this study reveals the interaction network among HuR, RRM2, and bifonazole and demonstrated that bifonazole is a potential therapeutic compound for ESCC through inhibition of the HuR/RRM2 axis.
Keywords: AU-rich elements (AREs); Bifonazole; Cell proliferation; Esophageal squamous cell carcinoma (ESCC); Hu antigen R (HuR); Ribonucleotide reductase small subunit M2 (RRM2); dNTP; mRNA stability.
© 2024 The Authors.
Conflict of interest statement
The authors declare no conflict of interests.
Figures



Similar articles
-
The Hu antigen R/interferon-β axis modulates sensitivity of esophageal squamous cancer cells to cisplatin.Neoplasma. 2022 Mar;69(2):383-391. doi: 10.4149/neo_2022_210910N1299. Epub 2022 Feb 28. Neoplasma. 2022. PMID: 35225647
-
SEMA4D under the posttranscriptional regulation of HuR and miR-4319 boosts cancer progression in esophageal squamous cell carcinoma.Cancer Biol Ther. 2020;21(2):122-129. doi: 10.1080/15384047.2019.1669996. Epub 2019 Oct 25. Cancer Biol Ther. 2020. PMID: 31651222 Free PMC article.
-
Gemcitabine resistance mediated by ribonucleotide reductase M2 in lung squamous cell carcinoma is reversed by GW8510 through autophagy induction.Clin Sci (Lond). 2018 Jul 9;132(13):1417-1433. doi: 10.1042/CS20180010. Print 2018 Jul 16. Clin Sci (Lond). 2018. PMID: 29853661
-
Ribonucleotide reductase subunit M2 as a novel target for clear-cell renal cell carcinoma.Onco Targets Ther. 2019 Apr 30;12:3267-3275. doi: 10.2147/OTT.S196347. eCollection 2019. Onco Targets Ther. 2019. PMID: 31118677 Free PMC article.
-
Ribonucleotide reductase M2 (RRM2): Regulation, function and targeting strategy in human cancer.Genes Dis. 2022 Dec 28;11(1):218-233. doi: 10.1016/j.gendis.2022.11.022. eCollection 2024 Jan. Genes Dis. 2022. PMID: 37588202 Free PMC article. Review.
Cited by
-
Construction and validation of a nomogram model for predicting peritoneal metastasis in gastric cancer based on ferroptosis-relate genes and clinicopathological features.J Gastrointest Oncol. 2025 Feb 28;16(1):264-280. doi: 10.21037/jgo-24-670. Epub 2025 Feb 26. J Gastrointest Oncol. 2025. PMID: 40115916 Free PMC article.
References
-
- Zhang G.C., Yu X.N., Guo H.Y., Sun J.L., Liu Z.Y., Zhu J.M., et al. PRP19 enhances esophageal squamous cell carcinoma progression by reprogramming SREBF1-dependent fatty acid metabolism. Cancer Res. 2023;83:521–537. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

