Whole genome sequencing data of Klebsiella pneumoniae Ch1-39 isolated from chili powder
- PMID: 39525660
- PMCID: PMC11546436
- DOI: 10.1016/j.dib.2024.111050
Whole genome sequencing data of Klebsiella pneumoniae Ch1-39 isolated from chili powder
Abstract
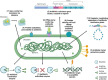
Klebsiella pneumoniae Ch1-39 was isolated from chili powder elaborated at San Luis Potosí, México. This microorganism can be found in diverse ecological niches as water, soil, air, plants and hospital setting, it is considered as a relevant opportunistic pathogen causing several diseases and showing increasingly multi-resistance to antibiotics. The genome was sequenced on the Illumina NovaSeq platform and bioinformatic analyses were made at the Bacterial and Viral Bioinformatics Resource Center (BV-BRC). The genome consisted of 72 contigs with a total size of 5,410,125 bp, 5,361 protein coding sequences (CDS), a total of 6 rRNA and 76 tRNA with an average G + C content of 57.22 %. The genome data was deposited at National Center for Biotechnology Information (NCBI) under accession number Bioproject ID PRJNA1062060, Bio Sample ID SAMN40269967. The genome accession number was JBAWUH000000000.
Keywords: Antibiotic Resistance genes (ARGs); Chili powder; Complete genome; Klebsiella pneumoniae Ch1-39; Virulence factor genes.
© 2024 The Author(s).
Figures


References
-
- Mena Navarro M.P., Espinosa Bernal M.A., Alvarado Osuna C., Ramos López M.Á., Amaro Reyes A., Arvizu Gómez J.L., Pacheco Aguilar J.R., Saldaña Gutiérrez C., Pérez Moreno V., Rodríguez Morales J.A., et al. A study of resistome in mexican chili powder as a public health risk factor. Antibiotics. 2024;13:182. doi: 10.3390/antibiotics13020182. - DOI - PMC - PubMed
-
- Olson R.D., Assaf R., Brettin T., Conrad N., Cucinell C., Davis J.J., Dempsey D.M., Dickerman A., Dietrich E.M., Kenyon R.W., et al. Introducing the bacterial and viral bioinformatics resource center (BV-BRC): a resource combining PATRIC, IRD and ViPR. Nucleic Acids Res. 2022;51:D678–D689. doi: 10.1093/nar/gkac1003. - DOI - PMC - PubMed
-
- Brettin T., Davis J.J., Disz T., Edwards R.A., Gerdes S., Olsen G.J., Olson R., Overbeek R., Parrello B., Pusch G.D., Shukla M., Thomason J.A., Stevens R., Vonstein V., Wattam A.R., Xia F. RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 2015;5:8365. doi: 10.1038/srep08365. - DOI - PMC - PubMed
-
- Cruickshank R. Churchill Livingstone; Edinburg: 1980. Medical Microbiology,12th eds. (revised reprint) pp. 170–189.
LinkOut - more resources
Full Text Sources

