PsPRE1 is a basic helix-loop-helix transcription factor that confers enhanced root growth and tolerance to salt stress in poplar
- PMID: 39526258
- PMCID: PMC11524248
- DOI: 10.48130/FR-2023-0016
PsPRE1 is a basic helix-loop-helix transcription factor that confers enhanced root growth and tolerance to salt stress in poplar
Abstract
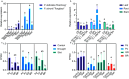
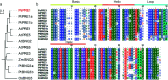
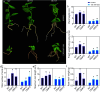
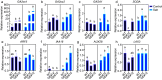
The basic helix-loop-helix (bHLH) family of transcription factors is one of the largest and oldest transcription factor families in plants. Members of the bHLH family regulate various growth and metabolic processes in plants. We used quantitative trait locus (QTL) mapping and transcriptome sequencing (RNA-seq) to identify PRE1 as a candidate bHLH transcription factor associated with root dry weight (RDW) in poplar. PRE1 was highly expressed in the roots and xylem, and was responsive to gibberellin, salicylic acid, drought, and salt stress. We cloned the PRE1 homolog from Populus simonii 'Tongliao1', referred to as PsPRE1, and transformed it into 84K poplar (Populus alba × Populus glandulosa). The overexpression of PsPRE1 in 84K poplar increased adventitious root development, fresh weight, total root number, and maximum root length. Poplar lines overexpressing PsPRE1 also exhibited enhanced salt tolerance while retaining a normal growth phenotype in the presence of salt stress. Catalase (CAT) activity in the PsPRE1 overexpression lines was higher than that of the wild-type, which may play a role in detoxifying stress-induced hydrogen peroxide production. An RNA-seq analysis of the PsPRE1 overexpression line revealed several differentially expressed genes (DEGs) involved in or related to auxin-, gibberellin-, and salicylic acid pathways, which indicates that the regulation of root development in poplar by PsPRE1 may involve multiple hormones.
Keywords: Adventitious root; Hormone; Poplar; PsPRE1; Quantitative trait locus; Salt stress.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

References
LinkOut - more resources
Full Text Sources
Miscellaneous
