Genome-wide identification and expression pattern analysis of the SABATH gene family in Neolamarckia cadamba
- PMID: 39526264
- PMCID: PMC11524262
- DOI: 10.48130/FR-2023-0013
Genome-wide identification and expression pattern analysis of the SABATH gene family in Neolamarckia cadamba
Abstract
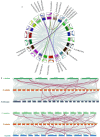
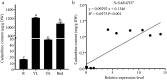
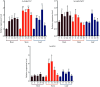
Plant SABATH methyltransferases are a class of enzymes that catalyze the transfer of the methyl group from S-adenosyl-L-methionine (SAM) to the carboxyl group or the nitrogen group of the substrate to form small molecule methyl esters or N-methylated compounds, which are involved in various secondary metabolite biosynthesis and have important impacts on plant growth, development, and defense reactions. We previously reported the monoterpenoid indole alkaloids (MIAs) cadambine biosynthetic pathway in Neolamarckia cadamba, a woody tree species that provides an important traditional medicine widely used to treat diseases such as diabetes, leprosy, and cancer in Southeast Asia. However, the functions of NcSABATHs in cadambine biosynthesis remain unclear. In this study, 23 NcSABATHs were identified and found to be distributed on 12 of the total 22 chromosomes. Gene structure, conserved motifs, and phylogenetic analysis showed that NcSABATHs could be divided into three groups. According to cis-element analysis, the NcSABATH promoters contained a large number of elements involved in light, plant hormone, and environmental stress responses, as well as binding sites for the BBR-BPC, DOF, and MYB transcription factor families. Based on RNA-seq data and qRT-PCR analysis, the NcSABATH genes exhibited diverse tissue expression patterns. Furthermore, NcSABATH7/22, which clustered with LAMT in the same clade, were both up-regulated under MeJA treatment. The correlation analysis between gene expression and cadambine content showed that NcSABATH7 potentially participated in cadambine biosynthesis. Taken together, our study not only enhanced our understanding of SABATH in N. cadamba but also identified potential candidate genes involved in cadambine biosynthesis.
Keywords: Cadambine; Expression pattern; Methyltransferase; Neolamarckia cadamba; SABATH gene family.
Conflict of interest statement
The authors declare that they have no conflict of interest. Changcao Peng is the Editorial Board member of Forestry Research, who was blinded from reviewing or making decisions on the manuscript. The article was subject to the journal's standard procedures, with peer-review handled independently of this Editorial Board member and his research groups.
Figures




References
-
- Ross JR, Nam KH, D’Auria JC, Pichersky E S-Adenosyl-L methionine: salicylic acid carboxyl methyltransferase, an enzyme involved in floral scent production and plant defense, represents a new class of plant methyltransferases. Archives of Biochemistry and Biophysics. 1999;367:9–16. doi: 10.1006/abbi.1999.1255. - DOI - PubMed
-
- Murfitt LM, Kolosova N, Mann CJ, Dudareva N Purification and characterization of S-adenosyl-l-methionine: benzoic acid carboxyl methyltransferase, the enzyme responsible for biosynthesis of the volatile ester methyl benzoate in flowers of Antirrhinum majus. Archives of Biochemistry and Biophysics. 2000;382:145–51. doi: 10.1006/abbi.2000.2008. - DOI - PubMed
LinkOut - more resources
Full Text Sources
