scProAtlas: an atlas of multiplexed single-cell spatial proteomics imaging in human tissues
- PMID: 39526396
- PMCID: PMC11701751
- DOI: 10.1093/nar/gkae990
scProAtlas: an atlas of multiplexed single-cell spatial proteomics imaging in human tissues
Abstract
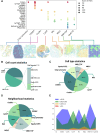
Spatial proteomics can visualize and quantify protein expression profiles within tissues at single-cell resolution. Although spatial proteomics can only detect a limited number of proteins compared to spatial transcriptomics, it provides comprehensive spatial information with single-cell resolution. By studying the spatial distribution of cells, we can clearly obtain the spatial context within tissues at multiple scales. Spatial context includes the spatial composition of cell types, the distribution of functional structures, and the spatial communication between functional regions, all of which are crucial for the patterns of cellular distribution. Here, we constructed a comprehensive spatial proteomics functional annotation knowledgebase, scProAtlas (https://relab.xidian.edu.cn/scProAtlas/#/), which is designed to help users comprehensively understand the spatial context within different tissue types at single-cell resolution and across multiple scales. scProAtlas contains multiple modules, including neighborhood analysis, proximity analysis and neighborhood network, to comprehensively construct spatial cell maps of tissues and multi-modal integration, spatial gene identification, cell-cell interaction and spatial pathway analysis to display spatial variable genes. scProAtlas includes data from eight spatial protein imaging techniques across 15 tissues and provides detailed functional annotation information for 17 468 394 cells from 945 region of interests. The aim of scProAtlas is to offer a new insight into the spatial structure of various tissues and provides detailed spatial functional annotation.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Chang Q., Ornatsky O.I., Siddiqui I., Loboda A., Baranov V.I., Hedley D.W.. Imaging mass cytometry. Cytometry A. 2017; 91:160–169. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

