Identification of hub genes in the crosstalk between type 2 diabetic nephropathy and obesity according to bioinformatics analysis
- PMID: 39526504
- PMCID: PMC11556279
- DOI: 10.1080/21623945.2024.2423723
Identification of hub genes in the crosstalk between type 2 diabetic nephropathy and obesity according to bioinformatics analysis
Abstract
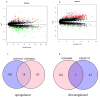
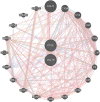
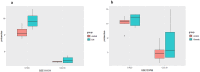
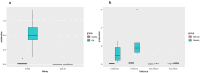
Diabetic nephropathy (DN) and obesity bring a huge burden to society. Obesity plays a crucial role in the progression of type 2 DN, but the pathophysiology remains unclear. Thus, we aimed the explore the association between type 2 DN and obesity using bioinformatics method. The gene expression profiles of type 2 DN (GSE96804) and obesity (GSE94752) were downloaded from the Gene Expression Omnibus (GEO) database. The differentially expressed genes (DEGs) were screened with the thresholds defined as |log2FC| ≥1 and P<0.05. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses were performed. Subsequently, a protein-protein interaction network was constructed based on the STRING database. Hub genes were identified, and the co-expression network was constructed. Finally, the hub genes were verified in clinical samples of 24 patients by immunohistochemistry. A total of 17 common DEGs were identified. Finally, two overlapping hub genes were identified (CCL18, C1QC). C1QC has been verified in clinical specimens. Using bioinformatics methods, the present study analyzed the common DEGs and the potential pathogenic mechanisms involved in type 2 DN and obesity. C1QC was the hub gene. Further studies are needed to clarify the specific relationships among C1QC, type 2 DN and obesity.
Keywords: Diabetic nephropathy; bioinformatics; differentially expressed genes; hub genes; obesity.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures



Similar articles
-
Investigation of the Mechanism of Complement System in Diabetic Nephropathy via Bioinformatics Analysis.J Diabetes Res. 2021 May 24;2021:5546199. doi: 10.1155/2021/5546199. eCollection 2021. J Diabetes Res. 2021. PMID: 34124269 Free PMC article.
-
Identification of Lipotoxicity-Related Biomarkers in Diabetic Nephropathy Based on Bioinformatic Analysis.J Diabetes Res. 2024 May 14;2024:5550812. doi: 10.1155/2024/5550812. eCollection 2024. J Diabetes Res. 2024. PMID: 38774257 Free PMC article.
-
Integrating bioinformatics and machine learning to identify glomerular injury genes and predict drug targets in diabetic nephropathy.Sci Rep. 2025 May 15;15(1):16868. doi: 10.1038/s41598-025-01628-5. Sci Rep. 2025. PMID: 40374840 Free PMC article.
-
Network pharmacology-based identification of miRNA expression of Astragalus membranaceus in the treatment of diabetic nephropathy.Medicine (Baltimore). 2022 Feb 4;101(5):e28747. doi: 10.1097/MD.0000000000028747. Medicine (Baltimore). 2022. PMID: 35119030 Free PMC article.
-
Elucidation of the molecular mechanism of type 2 diabetes mellitus affecting the progression of nonalcoholic steatohepatitis using bioinformatics and network pharmacology: A review.Medicine (Baltimore). 2024 Sep 13;103(37):e39731. doi: 10.1097/MD.0000000000039731. Medicine (Baltimore). 2024. PMID: 39287256 Free PMC article. Review.
Cited by
-
Exploring the potential molecular targets and therapeutic mechanisms of Cyperi Rhizoma in treating diabetic cardiomyopathy: a computational approach.RSC Adv. 2025 Jul 8;15(29):23920-23933. doi: 10.1039/d5ra02505b. eCollection 2025 Jul 4. RSC Adv. 2025. PMID: 40630680 Free PMC article.
References
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous
