Advancing Topoisomerase Research Using DNA Nanotechnology
- PMID: 39526512
- PMCID: PMC12182894
- DOI: 10.1002/smtd.202401113
Advancing Topoisomerase Research Using DNA Nanotechnology
Abstract
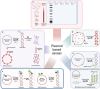
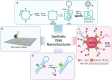
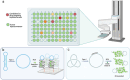
In this Perspective, the use of DNA nanotechnology is explored as a powerful tool for studying a family of enzymes known as topoisomerases. These enzymes regulate DNA topology within a living cell and play a major role in the pharmaceutical field, serving as anti-cancer and anti-bacterial targets. This Perspective will provide a short historical overview of the methods employed in studying these enzymes and emphasizing recent advancements in assays using DNA nanotechnology. These innovations have substantially improved accuracy and expanded the understanding of enzyme activity. This perspective will showcase the versatile utility of DNA nanotechnology in advancing scientific knowledge and its application in exploring new drug candidates, particularly in the study of topoisomerase enzymes.
Keywords: DNA nanotechnology; DNA topology; drug discovery; enzymatic assays; high‐thruput screening; topoisomerases.
© 2024 The Author(s). Small Methods published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
A Review of Natural and Synthetic Chalcones as Anticancer Agents Targeting Topoisomerase Enzymes.Molecules. 2025 Jun 6;30(12):2498. doi: 10.3390/molecules30122498. Molecules. 2025. PMID: 40572465 Free PMC article. Review.
-
Rapid detection of health-care-associated bloodstream infection in critical care using multipathogen real-time polymerase chain reaction technology: a diagnostic accuracy study and systematic review.Health Technol Assess. 2015 May;19(35):1-142. doi: 10.3310/hta19350. Health Technol Assess. 2015. PMID: 25961752 Free PMC article.
-
Development of a Marmoset Apparatus for Automated Pulling to study cooperative behaviors.Elife. 2024 Oct 28;13:RP97088. doi: 10.7554/eLife.97088. Elife. 2024. PMID: 39466838 Free PMC article.
-
Cost-effectiveness of using prognostic information to select women with breast cancer for adjuvant systemic therapy.Health Technol Assess. 2006 Sep;10(34):iii-iv, ix-xi, 1-204. doi: 10.3310/hta10340. Health Technol Assess. 2006. PMID: 16959170
-
Interventions to improve safe and effective medicines use by consumers: an overview of systematic reviews.Cochrane Database Syst Rev. 2014 Apr 29;2014(4):CD007768. doi: 10.1002/14651858.CD007768.pub3. Cochrane Database Syst Rev. 2014. PMID: 24777444 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

