Aberrant zonal recycling of germinal center B cells impairs appropriate selection in lupus
- PMID: 39527476
- PMCID: PMC11682828
- DOI: 10.1016/j.celrep.2024.114978
Aberrant zonal recycling of germinal center B cells impairs appropriate selection in lupus
Abstract
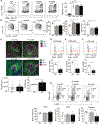
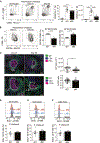
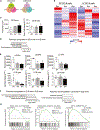
Autoimmune diseases such as lupus are characterized by polyclonal B cell activation, leading to the production of autoantibodies. The mechanism leading to B cell dysregulation is unclear; however, the defect may lie in selection within germinal centers (GCs). GC B cells cycle between proliferation and mutation in the dark zone and selection in the light zone (LZ). Temporal assessment of GCs from mice with either persistent infection or lupus showed an accumulation of LZ B cells. Yet, only in lupus, GC B cells exhibited reduced proliferation and progressive loss of MYC and FOXO1, which regulate zonal recycling and differentiation. As lupus progressed, decreased mutational frequency and repertoire diversity were associated with reduced responsiveness to CD40 signaling, despite accumulation of plasma cells. Collectively, these findings suggest that lupus disease progression coincides with an intrinsic defect in LZ B cell signaling, altering the zonal recycling, selection, and differentiation of autoreactive B cells.
Keywords: B cells; CP: Immunology; autoimmunity; germinal center.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors report no competing interests.
Figures




References
-
- MacLennan IC (1994). Germinal centers. Annu. Rev. Immunol. 12, 117–139. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

