STAMBPL1/TRIM21 Balances AXL Stability Impacting Mesenchymal Phenotype and Immune Response in KIRC
- PMID: 39527690
- PMCID: PMC11714167
- DOI: 10.1002/advs.202405083
STAMBPL1/TRIM21 Balances AXL Stability Impacting Mesenchymal Phenotype and Immune Response in KIRC
Abstract
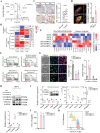
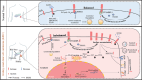
Kidney renal clear cell carcinoma (KIRC) is recognized as an immunogenic tumor, and immunotherapy is incorporated into its treatment landscape for decades. The acquisition of a tumor mesenchymal phenotype through epithelial-to-mesenchymal transition (EMT) is associated with immune evasion and can contribute to immunotherapy resistance. Here, the involvement of STAM Binding Protein Like 1 (STAMBPL1) is reported in the development of mesenchymal and immune evasion phenotypes in KIRC cells. Mechanistically, STAMBPL1 elevated protein abundance and surface accumulation of TAM Receptor AXL through diminishing the TRIM21-mediated K63-linked ubiquitination and subsequent lysosomal degradation of AXL, thereby enhancing the expression of mesenchymal genes while suppressing chemokines CXCL9/10 and HLA/B/C. In addition, STAMBPL1 enhanced PD-L1 transcription via facilitating nuclear translocation of p65, and knockdown (KD) of STAMBPL1 augmented antitumor effects of PD-1 blockade. Furthermore, STAMBPL1 silencing and the tyrosine kinase inhibitor (TKI) sunitinib also exhibited a synergistic effect on the suppression of KIRC. Collectively, targeting the STAMBPL1/TRIM21/AXL axis can decrease mesenchymal phenotype and potentiate anti-tumor efficacy of cancer therapy.
Keywords: STAM binding protein like 1; immunotherapy resistance; kidney renal clear cell carcinoma; mesenchymal phenotype; ubiquitination.
© 2024 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






Similar articles
-
Targeting MET and AXL overcomes resistance to sunitinib therapy in renal cell carcinoma.Oncogene. 2016 May;35(21):2687-97. doi: 10.1038/onc.2015.343. Epub 2015 Sep 14. Oncogene. 2016. PMID: 26364599 Free PMC article.
-
Clear Cell Renal Cell Carcinoma is linked to Epithelial-to-Mesenchymal Transition and to Fibrosis.Physiol Rep. 2017 Jun;5(11):e13305. doi: 10.14814/phy2.13305. Physiol Rep. 2017. PMID: 28596300 Free PMC article.
-
AXL targeting by a specific small molecule or monoclonal antibody inhibits renal cell carcinoma progression in an orthotopic mice model.Physiol Rep. 2021 Dec;9(23):e15140. doi: 10.14814/phy2.15140. Physiol Rep. 2021. PMID: 34877810 Free PMC article.
-
AXL-Driven EMT State as a Targetable Conduit in Cancer.Cancer Res. 2017 Jul 15;77(14):3725-3732. doi: 10.1158/0008-5472.CAN-17-0392. Epub 2017 Jun 30. Cancer Res. 2017. PMID: 28667075 Review.
-
Accelerating AXL targeting for TNBC therapy.Int J Biochem Cell Biol. 2021 Oct;139:106057. doi: 10.1016/j.biocel.2021.106057. Epub 2021 Aug 14. Int J Biochem Cell Biol. 2021. PMID: 34403827 Review.
Cited by
-
Genome-wide CRISPR/Cas9 screening identifies PTGR2 as a potential therapeutic target for sunitinib resistance in clear cell renal cell carcinoma.Sci Rep. 2025 Jul 19;15(1):26263. doi: 10.1038/s41598-025-12192-3. Sci Rep. 2025. PMID: 40683967 Free PMC article.
-
LPCAT3 regulates the immune infiltration and prognosis of ccRCC patients by mediating ferroptosis and endoplasmic reticulum stress.Discov Oncol. 2025 Apr 19;16(1):574. doi: 10.1007/s12672-025-02283-y. Discov Oncol. 2025. PMID: 40253575 Free PMC article.
References
-
- Diaz‐Montero C. M., Rini B. I., Finke J. H., Nat. Rev. Nephrol. 2020, 16, 721. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
