Adversarial regularized autoencoder graph neural network for microbe-disease associations prediction
- PMID: 39528423
- PMCID: PMC11554402
- DOI: 10.1093/bib/bbae584
Adversarial regularized autoencoder graph neural network for microbe-disease associations prediction
Abstract
Background: Microorganisms inhabit various regions of the human body and significantly contribute to numerous diseases. Predicting the associations between microbes and diseases is crucial for understanding pathogenic mechanisms and informing prevention and treatment strategies. Biological experiments to determine these associations are time-consuming and costly. Therefore, integrating deep learning with biological networks can efficiently identify potential microbe-disease associations on a large scale.
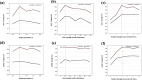
Methods: We propose an adversarial regularized autoencoder graph neural network algorithm, named Stacked Adversarial Regularization for Microbe-Disease Associations Prediction (SARMDA), for predicting associations between microbes and diseases. First, we integrate topological structural similarity and functional similarity metrics of microbes and diseases to construct a heterogeneous network. Then, utilizing an autoencoder based on GraphSAGE, we learn both the topological and attribute representations of nodes within the constructed network. Finally, we introduce an adversarial regularized autoencoder graph neural network embedding model to address the inherent limitations of traditional GraphSAGE autoencoders in capturing global information.
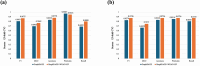
Results: Under the five-fold cross-validation on microbe-disease pairs, SARMDA was compared with eight advanced methods using the Human Microbe-Disease Association Database (HMDAD) and Disbiome databases. The best area under the ROC curve (AUC) achieved by SARMDA on HMDAD was 0.9891$\pm$0.0057, and the best area under the precision-recall curve (AUPR) was 0.9902$\pm$0.0128. On the Disbiome dataset, the AUC was 0.9328$\pm$0.0072, and the best AUPR was 0.9233$\pm$0.0089, outperforming the other eight MDAs prediction methods. Furthermore, the effectiveness of our model was demonstrated through a detailed analysis of asthma and inflammatory bowel disease cases.
Keywords: autoencoders; generative adversarial learning; graph neural networks; microbe-disease associations prediction.
© The Author(s) 2024. Published by Oxford University Press.
Figures



Similar articles
-
Predicting potential microbe-disease associations with graph attention autoencoder, positive-unlabeled learning, and deep neural network.Front Microbiol. 2023 Sep 18;14:1244527. doi: 10.3389/fmicb.2023.1244527. eCollection 2023. Front Microbiol. 2023. PMID: 37789848 Free PMC article.
-
MVGCNMDA: Multi-view Graph Augmentation Convolutional Network for Uncovering Disease-Related Microbes.Interdiscip Sci. 2022 Sep;14(3):669-682. doi: 10.1007/s12539-022-00514-2. Epub 2022 Apr 15. Interdiscip Sci. 2022. PMID: 35428964
-
Predicting potential microbe-disease associations based on dual branch graph convolutional network.J Cell Mol Med. 2024 Aug;28(15):e18571. doi: 10.1111/jcmm.18571. J Cell Mol Med. 2024. PMID: 39086148 Free PMC article.
-
MADGAN:A microbe-disease association prediction model based on generative adversarial networks.Front Microbiol. 2023 Mar 23;14:1159076. doi: 10.3389/fmicb.2023.1159076. eCollection 2023. Front Microbiol. 2023. PMID: 37032881 Free PMC article.
-
Genetically and semantically aware homogeneous network for prediction and scoring of comorbidities.Comput Biol Med. 2024 Dec;183:109252. doi: 10.1016/j.compbiomed.2024.109252. Epub 2024 Oct 16. Comput Biol Med. 2024. PMID: 39418770 Review.
Cited by
-
Harnessing dual variational autoencoders to decode microbe roles in diseases for traditional medicine discovery.Front Pharmacol. 2025 May 30;16:1578140. doi: 10.3389/fphar.2025.1578140. eCollection 2025. Front Pharmacol. 2025. PMID: 40520163 Free PMC article.
References
-
- Mazmanian SK, Liu CH, Tzianabos AO. et al. . An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 2005;122:107–18. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

