Integrative ensemble modelling of cetuximab sensitivity in colorectal cancer patient-derived xenografts
- PMID: 39528460
- PMCID: PMC11555063
- DOI: 10.1038/s41467-024-53163-y
Integrative ensemble modelling of cetuximab sensitivity in colorectal cancer patient-derived xenografts
Abstract
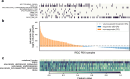
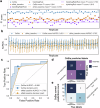
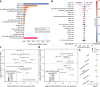
Patient-derived xenografts (PDXs) are tumour fragments engrafted into mice for preclinical studies. PDXs offer clear advantages over simpler in vitro cancer models - such as cancer cell lines (CCLs) and organoids - in terms of structural complexity, heterogeneity, and stromal interactions. Here, we characterise 231 colorectal cancer PDXs at the genomic, transcriptomic, and epigenetic levels, along with their response to cetuximab, an EGFR inhibitor used clinically for metastatic colorectal cancer. After evaluating the PDXs' quality, stability, and molecular concordance with publicly available patient cohorts, we present results from training, interpreting, and validating the integrative ensemble classifier CeSta. This model takes in input the PDXs' multi-omic characterisation and predicts their sensitivity to cetuximab treatment, achieving an area under the receiver operating characteristics curve > 0.88. Our study demonstrates that large PDX collections can be leveraged to train accurate, interpretable drug sensitivity models that: (1) better capture patient-derived therapeutic biomarkers compared to models trained on CCL data, (2) can be robustly validated across independent PDX cohorts, and (3) could contribute to the development of future therapeutic biomarkers.
© 2024. The Author(s).
Conflict of interest statement
Figures

References
-
- Biller, L. H. & Schrag, D. Diagnosis and treatment of metastatic colorectal cancer: a review. JAMA325, 669–685 (2021). - PubMed
-
- Bertotti, A. et al. A molecularly annotated platform of patient-derived xenografts (‘xenopatients’) identifies HER2 as an effective therapeutic target in cetuximab-resistant colorectal cancer. Cancer Discov.1, 508–523 (2011). - PubMed
-
- Stintzing, S. et al. FOLFIRI plus cetuximab versus FOLFIRI plus bevacizumab for metastatic colorectal cancer (FIRE-3): a post-hoc analysis of tumour dynamics in the final RAS wild-type subgroup of this randomised open-label phase 3 trial. Lancet Oncol.17, 1426–1434 (2016). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 28772/Associazione Italiana per la Ricerca sul Cancro (Italian Association for Cancer Research)
- 22802/Associazione Italiana per la Ricerca sul Cancro (Italian Association for Cancer Research)
- 22795/Associazione Italiana per la Ricerca sul Cancro (Italian Association for Cancer Research)
- 20697/Associazione Italiana per la Ricerca sul Cancro (Italian Association for Cancer Research)
- 21091/Associazione Italiana per la Ricerca sul Cancro (Italian Association for Cancer Research)
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

