Single-cell landscape identified SERPINB9 as a key player contributing to stemness and metastasis in non-seminomas
- PMID: 39528470
- PMCID: PMC11555415
- DOI: 10.1038/s41419-024-07220-5
Single-cell landscape identified SERPINB9 as a key player contributing to stemness and metastasis in non-seminomas
Abstract
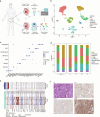
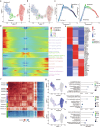
Embryonal carcinoma (EC), characterized by a high degree of stemness similar to that of embryonic stem cells, is the most malignant subtype within non-seminomatous testicular germ cell tumors (TGCTs). However, the mechanisms underlying its malignancy remain unknown. In this study, we employed single-cell RNA sequencing to analyze four non-seminoma samples. Our differential expression analysis revealed high expression of SERPINB9 in metastatic EC cells. We conducted in vitro experiments to further investigate SERPINB9's role in the progression of EC. Functionally, the knockdown of SERPINB9 in NCCIT and NTERA-2 leads to a diminished migratory capability and decreased cis-platin resistance, as demonstrated by Transwell migration assay and drug sensitivity assay. Moreover, embryoid bodies showed reduced size and lower OCT4 expression, alongside heightened expression of differentiation markers AFP, ACTA2, and CD57 in shSERPINB9 cells. In vivo, the role of SERPINB9 in maintaining cancer stemness was validated by the limiting dilution assay. Mechanistically, Bulk RNA-seq further showed downregulation of ERK1/2 signaling and WNT signaling pathways with concomitant upregulation of differentiation pathways subsequent to SERPINB9 knockdown. Additionally, the analysis indicated increased levels of cytokines linked to tertiary lymphoid structures (TLS), such as IL6, IL11, IL15, CCL2, CCL5, and CXCL13 in shSERPINB9 cells, which were further validated by ELISA. Our research indicates that SERPINB9 plays a key role in driving tumor progression by enhancing tumor stemness and suppressing TLS. This study stands as the first to elucidate the molecular signature of non-seminomas at a single-cell level, presenting a wealth of promising targets with substantial potential for informing the development of future therapeutic interventions.
© 2024. The Author(s).
Conflict of interest statement
Figures



Similar articles
-
Vitamin D metabolism and effects on pluripotency genes and cell differentiation in testicular germ cell tumors in vitro and in vivo.Neoplasia. 2012 Oct;14(10):952-63. doi: 10.1593/neo.121164. Neoplasia. 2012. PMID: 23097629 Free PMC article.
-
Overexpression of SerpinB9 in non-seminomatous germ cell tumors.Med Mol Morphol. 2024 Mar;57(1):68-75. doi: 10.1007/s00795-023-00374-9. Epub 2023 Nov 22. Med Mol Morphol. 2024. PMID: 37991604
-
Loss of Oct-3/4 expression in embryonal carcinoma cells is associated with induction of cisplatin resistance.Tumour Biol. 2006;27(2):71-83. doi: 10.1159/000092324. Epub 2006 Mar 24. Tumour Biol. 2006. PMID: 16557044
-
Chromosomes and expression in human testicular germ-cell tumors: insight into their cell of origin and pathogenesis.Ann N Y Acad Sci. 2007 Dec;1120:187-214. doi: 10.1196/annals.1411.000. Epub 2007 Oct 2. Ann N Y Acad Sci. 2007. PMID: 17911410 Review.
-
The role of microenvironment in testicular germ cell tumors.Cancer Biol Ther. 2010 Sep 15;10(6):529-36. doi: 10.4161/cbt.10.6.13227. Epub 2010 Sep 3. Cancer Biol Ther. 2010. PMID: 20855948 Review.
Cited by
-
Multiomics Approach Distinguishes SPTBN4 as a Key Molecule in Diagnosis, Prognosis, and Immune Suppression of Testicular Seminomas.Int J Genomics. 2025 Apr 25;2025:3530098. doi: 10.1155/ijog/3530098. eCollection 2025. Int J Genomics. 2025. PMID: 40321316 Free PMC article.
-
A Rare Case of Mediastinal Seminoma with Granulomatous Features: Diagnostic and Therapeutic Considerations.Surg Case Rep. 2025;11(1):25-0091. doi: 10.70352/scrj.cr.25-0091. Epub 2025 Jul 1. Surg Case Rep. 2025. PMID: 40612978 Free PMC article.
References
-
- Bosl GJ, Motzer RJ. Testicular germ-cell cancer. N. Engl J Med. 1997;337:242–54. - PubMed
-
- Germà-Lluch JR, Garcia del Muro X, Maroto P, Paz-Ares L, Arranz JA, Gumà J, et al. Clinical Pattern and Therapeutic Results Achieved in 1490 Patients with Germ-Cell Tumours of the Testis: the Experience of the Spanish Germ-Cell Cancer Group (GG. Eur Urol. 2002;42:553–63. - PubMed
-
- Vergouwe Y, Steyerberg EW, Eijkemans MJC, Albers P, Habbema JDF. Predictors of occult metastasis in clinical stage I nonseminoma: A systematic review. J Clin Oncol. 2003;21:4092–9. - PubMed
-
- Hermans BP, Sweeney CJ, Foster RS, Einhorn LE, Donohue JP. RISK OF SYSTEMIC METASTASES IN CLINICAL STAGE I NONSEMINOMA GERM CELL TESTIS TUMOR MANAGED BY RETROPERITONEAL LYMPH NODE DISSECTION. J Urol. 2000;163:1721–4. - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

