Chronic social stress induces p16-mediated senescent cell accumulation in mice
- PMID: 39528642
- PMCID: PMC12240448
- DOI: 10.1038/s43587-024-00743-8
Chronic social stress induces p16-mediated senescent cell accumulation in mice
Abstract
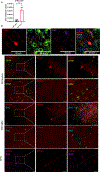
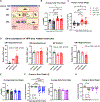
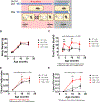
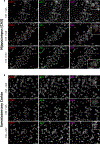
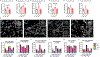
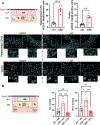
Life stress can shorten lifespan and increase risk for aging-related diseases, but the biology underlying this phenomenon remains unclear. Here we assessed the effect of chronic stress on cellular senescence-a hallmark of aging. Exposure to restraint stress, a psychological non-social stress model, increased p21Cip1 exclusively in the brains of male, but not female mice, and in a p16Ink4a-independent manner. Conversely, exposure to chronic subordination stress (only males were tested) increased key senescent cell markers in peripheral blood mononuclear cells, adipose tissue and brain, in a p16Ink4a-dependent manner. p16Ink4a-positive cells in the brain of chronic subordination stress-exposed mice were primarily hippocampal and cortical neurons with evidence of DNA damage that could be reduced by p16Ink4a cell clearance. Clearance of p16Ink4a-positive cells was not sufficient to ameliorate the adverse effects of social stress on measured metrics of healthspan. Overall, our findings indicate that social stress induces an organ-specific and p16Ink4a-dependent accumulation of senescent cells, illuminating a fundamental way by which the social environment can contribute to aging.
© 2024. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing interests: D.J.B. and J.M.v.D. have a potential financial interest related to this research. J.M.v.D. is a cofounder of Unity Biotechnology, D.J.B. and J.M.v.D. are co-inventors on patents held by Mayo Clinic and patent applications licensed to or filed by Unity Biotechnology, and D.J.B. and J.M.v.D. are Unity shareholders. Research in the Baker laboratory has been reviewed by the Mayo Clinic Conflict of Interest Review Board and is being conducted in compliance with Mayo Clinic Conflict of Interest policies. The other authors declare no conflicts of interest.
Figures
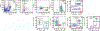









References
-
- Gu HF, Tang CK & Yang YZ Psychological stress, immune response, and atherosclerosis. Atherosclerosis 223, 69–77 (2012). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

