Proteogenomic analysis of human cerebrospinal fluid identifies neurologically relevant regulation and implicates causal proteins for Alzheimer's disease
- PMID: 39528825
- PMCID: PMC11831731
- DOI: 10.1038/s41588-024-01972-8
Proteogenomic analysis of human cerebrospinal fluid identifies neurologically relevant regulation and implicates causal proteins for Alzheimer's disease
Abstract
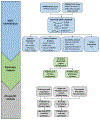
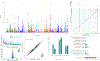
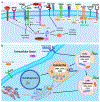
The integration of quantitative trait loci (QTLs) with disease genome-wide association studies (GWASs) has proven successful in prioritizing candidate genes at disease-associated loci. QTL mapping has been focused on multi-tissue expression QTLs or plasma protein QTLs (pQTLs). We generated a cerebrospinal fluid (CSF) pQTL atlas by measuring 6,361 proteins in 3,506 samples. We identified 3,885 associations for 1,883 proteins, including 2,885 new pQTLs, demonstrating unique genetic regulation in CSF. We identified CSF-enriched pleiotropic regions on chromosome (chr)3q28 near OSTN and chr19q13.32 near APOE that were enriched for neuron specificity and neurological development. We integrated our associations with Alzheimer's disease (AD) through proteome-wide association study (PWAS), colocalization and Mendelian randomization and identified 38 putative causal proteins, 15 of which have drugs available. Finally, we developed a proteomics-based AD prediction model that outperforms genetics-based models. These findings will be instrumental to further understand the biology and identify causal and druggable proteins for brain and neurological traits.
© 2024. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing interests: C.C. has received research support from GSK and Eisai. The funders of the study had no role in the collection, analysis or interpretation of data; in the writing of the report; or in the decision to submit the paper for publication. C.C. is a member of the advisory board of Circular Genomics and owns stocks in this company. D.J.P. is an employee of GSK and holds stock in GSK. M.d.C.M. has been an invited speaker at Eisai. M.d.C.M. is an associate editor at Alzheimer’s Research and Therapy. B.T. and P.J.V. are inventors on a patent (WO2020197399A1, owned by Stichting VUmc). C.E.T. has a collaboration contract with ADx Neurosciences, Quanterix and Eli Lilly and performed contract research or received grants from AC Immune, Axon Neuroscience, BioConnect, Bioorchestra, Brainstorm Therapeutics, Celgene, EIP Pharma, Eisai, Grifols, Novo Nordisk, PeopleBio, Roche, Toyama and Vivoryon. She serves on editorial boards of Medidact Neurologie–Springer, Alzheimer’s Research and Therapy and Neurology: Neuroimmunology and Neuroinflammation and is an editor of the Neuromethods book (Springer). She had speaker contracts for Roche, Grifols and Novo Nordisk. The rest of the authors declare no competing interest.
Figures

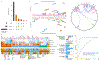
Update of
-
Proteogenomic analysis of human cerebrospinal fluid identifies neurologically relevant regulation and informs causal proteins for Alzheimer's disease.Res Sq [Preprint]. 2023 Jun 9:rs.3.rs-2814616. doi: 10.21203/rs.3.rs-2814616/v1. Res Sq. 2023. Update in: Nat Genet. 2024 Dec;56(12):2672-2684. doi: 10.1038/s41588-024-01972-8. PMID: 37333337 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
- P30 AG066444/AG/NIA NIH HHS/United States
- RF1 AG071706/AG/NIA NIH HHS/United States
- U01 AG024904/AG/NIA NIH HHS/United States
- RF1 AG074007/AG/NIA NIH HHS/United States
- RF1AG058501/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- RF1AG053303/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- R01 AG074007/AG/NIA NIH HHS/United States
- R01AG044546/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- U19 AG032438/AG/NIA NIH HHS/United States
- R01 AG058501/AG/NIA NIH HHS/United States
- U01AG058922/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- U01 AG058922/AG/NIA NIH HHS/United States
- R01 AG044546/AG/NIA NIH HHS/United States
- P01AG003991/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- P30 AG066515/AG/NIA NIH HHS/United States
- RF1 AG053303/AG/NIA NIH HHS/United States
- R01 AG078964/AG/NIA NIH HHS/United States
- P01 AG003991/AG/NIA NIH HHS/United States
- P01 AG026276/AG/NIA NIH HHS/United States
- RF1 AG058501/AG/NIA NIH HHS/United States
- ZEN-22-848604/ALZ/Alzheimer's Association/United States
- R00 AG062723/AG/NIA NIH HHS/United States
- R01 AG064614/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

