Comparative transcriptomic analyses of thymocytes using 10x Genomics and Parse scRNA-seq technologies
- PMID: 39528918
- PMCID: PMC11552371
- DOI: 10.1186/s12864-024-10976-x
Comparative transcriptomic analyses of thymocytes using 10x Genomics and Parse scRNA-seq technologies
Abstract
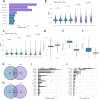
Background: Single-cell RNA sequencing experiments commonly use 10x Genomics (10x) kits due to their high-throughput capacity and standardized protocols. Recently, Parse Biosciences (Parse) introduced an alternative technology that uses multiple in-situ barcoding rounds within standard 96-well plates. Parse enables the analysis of more cells from multiple samples in a single run without the need for additional reagents or specialized microfluidics equipment. To evaluate the performance of both platforms, we conducted a benchmark study using biological and technical replicates of mouse thymus as a complex immune tissue.
Results: We found that Parse detected nearly twice the number of genes compared to 10x, with each platform detecting a distinct set of genes. The comparison of multiplexed samples generated from 10x and Parse techniques showed 10x data to have lower technical variability and more precise annotation of biological states in the thymus compared to Parse.
Conclusion: Our results provide a comprehensive comparison of the suitability of both single-cell platforms for immunological studies.
Keywords: 10x; Parse; Thymus; Transcriptomics; scRNA-seq.
© 2024. The Author(s).
Conflict of interest statement
Figures




Similar articles
-
Direct Comparative Analyses of 10X Genomics Chromium and Smart-seq2.Genomics Proteomics Bioinformatics. 2021 Apr;19(2):253-266. doi: 10.1016/j.gpb.2020.02.005. Epub 2021 Mar 2. Genomics Proteomics Bioinformatics. 2021. PMID: 33662621 Free PMC article.
-
Comparative Analysis of Single-Cell RNA Sequencing Methods with and without Sample Multiplexing.Int J Mol Sci. 2024 Mar 29;25(7):3828. doi: 10.3390/ijms25073828. Int J Mol Sci. 2024. PMID: 38612639 Free PMC article.
-
Single-Cell Transcriptomics of Immune Cells: Cell Isolation and cDNA Library Generation for scRNA-Seq.Methods Mol Biol. 2020;2184:1-18. doi: 10.1007/978-1-0716-0802-9_1. Methods Mol Biol. 2020. PMID: 32808214
-
A Single-Cell Sequencing Guide for Immunologists.Front Immunol. 2018 Oct 23;9:2425. doi: 10.3389/fimmu.2018.02425. eCollection 2018. Front Immunol. 2018. PMID: 30405621 Free PMC article. Review.
-
Data normalization for addressing the challenges in the analysis of single-cell transcriptomic datasets.BMC Genomics. 2024 May 6;25(1):444. doi: 10.1186/s12864-024-10364-5. BMC Genomics. 2024. PMID: 38711017 Free PMC article. Review.
References
-
- Zeng Y, Liu C, Gong Y, Bai Z, Hou S, He J, et al. Single-Cell RNA Sequencing Resolves Spatiotemporal Development of Pre-thymic Lymphoid Progenitors and Thymus Organogenesis in Human Embryos. Immunity. 2019;51(5):930-948.e6. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

