An integrated microbiome- and metabolome-genome-wide association study reveals the role of heritable ruminal microbial carbohydrate metabolism in lactation performance in Holstein dairy cows
- PMID: 39529146
- PMCID: PMC11555892
- DOI: 10.1186/s40168-024-01937-3
An integrated microbiome- and metabolome-genome-wide association study reveals the role of heritable ruminal microbial carbohydrate metabolism in lactation performance in Holstein dairy cows
Erratum in
-
Correction: An integrated microbiome- and metabolome-genome-wide association study reveals the role of heritable ruminal microbial carbohydrate metabolism in lactation performance in Holstein dairy cows.Microbiome. 2025 Feb 7;13(1):47. doi: 10.1186/s40168-025-02040-x. Microbiome. 2025. PMID: 39920859 Free PMC article. No abstract available.
Abstract
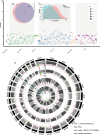
Background: Despite the growing number of studies investigating the connection between host genetics and the rumen microbiota, there remains a dearth of systematic research exploring the composition, function, and metabolic traits of highly heritable rumen microbiota influenced by host genetics. Furthermore, the impact of these highly heritable subsets on lactation performance in cows remains unknown. To address this gap, we collected and analyzed whole-genome resequencing data, rumen metagenomes, rumen metabolomes and short-chain fatty acids (SCFAs) content, and lactation performance phenotypes from a cohort of 304 dairy cows.
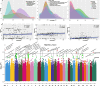
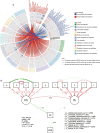
Results: The results indicated that the proportions of highly heritable subsets (h2 ≥ 0.2) of the rumen microbial composition (55%), function (39% KEGG and 28% CAZy), and metabolites (18%) decreased sequentially. Moreover, the highly heritable microbes can increase energy-corrected milk (ECM) production by reducing the rumen acetate/propionate ratio, according to the structural equation model (SEM) analysis (CFI = 0.898). Furthermore, the highly heritable enzymes involved in the SCFA synthesis metabolic pathway can promote the synthesis of propionate and inhibit the acetate synthesis. Next, the same significant SNP variants were used to integrate information from genome-wide association studies (GWASs), microbiome-GWASs, metabolome-GWASs, and microbiome-wide association studies (mWASs). The identified single nucleotide polymorphisms (SNPs) of rs43470227 and rs43472732 on SLC30A9 (Zn2+ transport) (P < 0.05/nSNPs) can affect the abundance of rumen microbes such as Prevotella_sp., Prevotella_sp._E15-22, Prevotella_sp._E13-27, which have the oligosaccharide-degradation enzymes genes, including the GH10, GH13, GH43, GH95, and GH115 families. The identified SNPs of chr25:11,177 on 5s_rRNA (small ribosomal RNA) (P < 0.05/nSNPs) were linked to ECM, the abundance alteration of Pseudobutyrivibrio_sp. (a genus that was also showed to be linked to the ECM production via the mWASs analysis), GH24 (lysozyme), and 9,10,13-TriHOME (linoleic acid metabolism). Moreover, ECM, and the abundances of Pseudobutyrivibrio sp., GH24, and 9,10,13-TRIHOME were significantly greater in the GG genotype than in the AG genotype at chr25:11,177 (P < 0.05). By further the SEM analysis, GH24 was positively correlated with Pseudobutyrivibrio sp., which was positively correlated with 9,10,13-triHOME and subsequently positively correlated with ECM (CFI = 0.942).
Conclusion: Our comprehensive study revealed the distinct heritability patterns of rumen microbial composition, function, and metabolism. Additionally, we shed light on the influence of host SNP variants on the rumen microbes with carbohydrate metabolism and their subsequent effects on lactation performance. Collectively, these findings offer compelling evidence for the host-microbe interactions, wherein cows actively modulate their rumen microbiota through SNP variants to regulate their own lactation performance. Video Abstract.
Keywords: Dairy cow; GWAS; Heritability; Host genetics; Lactation performance; Ruminal metabolome; Ruminal metagenome.
© 2024. The Author(s).
Conflict of interest statement
Figures



References
-
- FAO. Food Outlook Biannual report on global food markets. Rome: Food and Agriculture Organization of the United Nations; 2022. 10.4060/cb9427en.
-
- Rask KJ, Rask N. Economic development and food production–consumption balance: a growing global challenge. Food Policy. 2011;36(2):186–96. 10.1016/j.foodpol.2010.11.015.
-
- Tricarico JM, Kebreab E, Wattiaux MA. MILK symposium review: sustainability of dairy production and consumption in low-income countries with emphasis on productivity and environmental impact*. J Dairy Sci. 2020;103(11):9791–802. 10.3168/jds.2020-18269. - PubMed
-
- Schennink A, Stoop WM, Visker MHPW, Heck JML, Bovenhuis H, Van Der Poel JJ, Van Valenberg HJF, Van Arendonk JAM. DGAT1 underlies large genetic variation in milk-fat composition of dairy cows. Animal Genet. 2007;38(5):467–73. 10.1111/j.1365-2052.2007.01635.x. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

