BCR, not TCR, repertoire diversity is associated with favorable COVID-19 prognosis
- PMID: 39530088
- PMCID: PMC11550956
- DOI: 10.3389/fimmu.2024.1405013
BCR, not TCR, repertoire diversity is associated with favorable COVID-19 prognosis
Abstract
Introduction: The SARS-CoV-2 pandemic has had a widespread and severe impact on society, yet there have also been instances of remarkable recovery, even in critically ill patients.
Materials and methods: In this study, we used single-cell RNA sequencing to analyze the immune responses in recovered and deceased COVID-19 patients during moderate and critical stages.
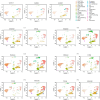
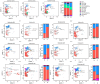
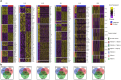
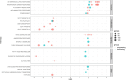
Results: Expanded T cell receptor (TCR) clones were predominantly SARS-CoV-2-specific, but represented only a small fraction of the total repertoire in all patients. In contrast, while deceased patients exhibited monoclonal B cell receptor (BCR) expansions without COVID-19 specificity, survivors demonstrated diverse and specific BCR clones. These findings suggest that neither TCR diversity nor BCR monoclonal expansions are sufficient for viral clearance and subsequent recovery. Differential gene expression analysis revealed that protein biosynthetic processes were enriched in survivors, but that potentially damaging mitochondrial ATP metabolism was activated in the deceased.
Conclusion: This study underscores that BCR repertoire diversity, but not TCR diversity, correlates with favorable outcomes in COVID-19.
Keywords: COVID - 19; gene expression; immune repertoire analysis; immunology & infectious diseases; single cell RNA and transcriptome sequencing.
Copyright © 2024 Paran, Oyama, Khasawneh, Ai, Ismanto, Sherif, Saputri, Ono, Saita, Takei, Horiuchi, Yagi, Matsuura, Okazaki, Takahashi, Standley, Tabe and Naito.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

